Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000375A_C01 KMC000375A_c01
(559 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
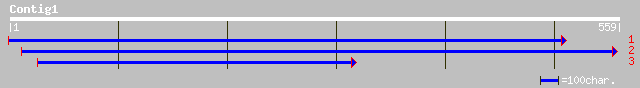
dbj|BAB08509.1| DNA excision repair cross-complementing protein;... 104 8e-22
pir||S71206 DNA excision repair cross-complementing protein ERCC... 104 8e-22
ref|NP_568592.1| TFIIH basal transcription factor complex helica... 104 8e-22
dbj|BAB08508.1| DNA excision repair cross-complementing protein ... 95 7e-19
ref|NP_568591.1| TFIIH basal transcription factor complex helica... 95 7e-19
>dbj|BAB08509.1| DNA excision repair cross-complementing protein; similar to human
Xeroderma pigmentosum group B DNA repair protein
[Arabidopsis thaliana]
Length = 755
Score = 104 bits (259), Expect = 8e-22
Identities = 51/73 (69%), Positives = 58/73 (78%)
Frame = -3
Query: 557 SAGDDAVGLEQLEEDTDEIALNSARRSHGSMSAMSGAKGMFYMEYSTGRKGQGQIKSKPK 378
+AGDD VGLEQLEEDTD +AL ARRS GSMS MSG+KGM YMEY++GR GQ KPK
Sbjct: 682 NAGDDLVGLEQLEEDTDGMALQKARRSMGSMSVMSGSKGMVYMEYNSGRHKSGQQFKKPK 741
Query: 377 DPAKRHFLFKRRF 339
DP KRH LFK+R+
Sbjct: 742 DPTKRHNLFKKRY 754
>pir||S71206 DNA excision repair cross-complementing protein ERCC3 - Arabidopsis
thaliana
Length = 757
Score = 104 bits (259), Expect = 8e-22
Identities = 51/73 (69%), Positives = 58/73 (78%)
Frame = -3
Query: 557 SAGDDAVGLEQLEEDTDEIALNSARRSHGSMSAMSGAKGMFYMEYSTGRKGQGQIKSKPK 378
+AGDD VGLEQLEEDTD +AL ARRS GSMS MSG+KGM YMEY++GR GQ KPK
Sbjct: 684 NAGDDLVGLEQLEEDTDGMALQKARRSMGSMSVMSGSKGMVYMEYNSGRHKSGQQFKKPK 743
Query: 377 DPAKRHFLFKRRF 339
DP KRH LFK+R+
Sbjct: 744 DPTKRHNLFKKRY 756
>ref|NP_568592.1| TFIIH basal transcription factor complex helicase XPB subunit,
putative; protein id: At5g41370.1, supported by cDNA:
gi_10314019, supported by cDNA: gi_14517423 [Arabidopsis
thaliana] gi|11771735|gb|AAC49987.2| putative DNA repair
protein and transcription factor [Arabidopsis thaliana]
gi|14517424|gb|AAK62602.1| AT5g41370/MYC6_8 [Arabidopsis
thaliana] gi|21360401|gb|AAM47316.1| AT5g41370/MYC6_8
[Arabidopsis thaliana]
Length = 767
Score = 104 bits (259), Expect = 8e-22
Identities = 51/73 (69%), Positives = 58/73 (78%)
Frame = -3
Query: 557 SAGDDAVGLEQLEEDTDEIALNSARRSHGSMSAMSGAKGMFYMEYSTGRKGQGQIKSKPK 378
+AGDD VGLEQLEEDTD +AL ARRS GSMS MSG+KGM YMEY++GR GQ KPK
Sbjct: 694 NAGDDLVGLEQLEEDTDGMALQKARRSMGSMSVMSGSKGMVYMEYNSGRHKSGQQFKKPK 753
Query: 377 DPAKRHFLFKRRF 339
DP KRH LFK+R+
Sbjct: 754 DPTKRHNLFKKRY 766
>dbj|BAB08508.1| DNA excision repair cross-complementing protein [Arabidopsis
thaliana]
Length = 754
Score = 94.7 bits (234), Expect = 7e-19
Identities = 48/73 (65%), Positives = 56/73 (75%)
Frame = -3
Query: 557 SAGDDAVGLEQLEEDTDEIALNSARRSHGSMSAMSGAKGMFYMEYSTGRKGQGQIKSKPK 378
+AGDD VGLEQLEEDTD AL + RRS GSMSAMSGA G YMEY++GR+ G KPK
Sbjct: 682 NAGDDMVGLEQLEEDTDGKALKT-RRSMGSMSAMSGANGRVYMEYNSGRQKSGNQSKKPK 740
Query: 377 DPAKRHFLFKRRF 339
DP KRH +FK+R+
Sbjct: 741 DPTKRHNIFKKRY 753
>ref|NP_568591.1| TFIIH basal transcription factor complex helicase XPB subunit,
putative; protein id: At5g41360.1, supported by cDNA:
gi_11037021 [Arabidopsis thaliana]
gi|11037022|gb|AAG27465.1|AF308595_1 putative DNA repair
protein and transcription factor [Arabidopsis thaliana]
Length = 766
Score = 94.7 bits (234), Expect = 7e-19
Identities = 48/73 (65%), Positives = 56/73 (75%)
Frame = -3
Query: 557 SAGDDAVGLEQLEEDTDEIALNSARRSHGSMSAMSGAKGMFYMEYSTGRKGQGQIKSKPK 378
+AGDD VGLEQLEEDTD AL + RRS GSMSAMSGA G YMEY++GR+ G KPK
Sbjct: 694 NAGDDMVGLEQLEEDTDGKALKT-RRSMGSMSAMSGANGRVYMEYNSGRQKSGNQSKKPK 752
Query: 377 DPAKRHFLFKRRF 339
DP KRH +FK+R+
Sbjct: 753 DPTKRHNIFKKRY 765
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 462,721,894
Number of Sequences: 1393205
Number of extensions: 9502553
Number of successful extensions: 22382
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 21615
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22366
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)