Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000372A_C01 KMC000372A_c01
(673 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
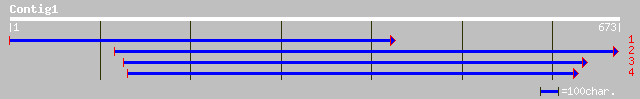
gb|AAD46410.1|AF096260_1 ER66 protein [Lycopersicon esculentum] 166 3e-40
gb|AAG39222.1|AF253511_1 anther ethylene-upregulated protein ER1... 162 5e-39
pir||A84611 hypothetical protein At2g22300 [imported] - Arabidop... 159 3e-38
ref|NP_179817.2| ethylene-induced calmodulin-binding protein, pu... 159 3e-38
gb|AAN74651.1| calmodulin-binding transcription factor SR1 [Arab... 159 3e-38
>gb|AAD46410.1|AF096260_1 ER66 protein [Lycopersicon esculentum]
Length = 558
Score = 166 bits (419), Expect = 3e-40
Identities = 81/109 (74%), Positives = 92/109 (84%)
Frame = -1
Query: 673 KIQAHVRGHQVRKNHGKIIWSVGILEKVVLRWRRKGCGLRGFKSEGVSEGNMVQGESSTE 494
KIQAHVRGHQVR + IIWSVGILEKV+LRWRRKG GLRGFK E +EG+ +Q + E
Sbjct: 413 KIQAHVRGHQVRNKYKNIIWSVGILEKVILRWRRKGSGLRGFKPEAPTEGSNMQDQPVQE 472
Query: 493 DEYDLLKEGRKQTEQRLQIALARVKSMVQYPEARDQYHRLLNVVTEIQE 347
D+YD LKEGRKQTE+RLQ AL RVKSMVQYPEARDQY RLLNVV+++QE
Sbjct: 473 DDYDFLKEGRKQTEERLQKALERVKSMVQYPEARDQYRRLLNVVSDMQE 521
>gb|AAG39222.1|AF253511_1 anther ethylene-upregulated protein ER1 [Nicotiana tabacum]
Length = 672
Score = 162 bits (409), Expect = 5e-39
Identities = 81/110 (73%), Positives = 93/110 (83%), Gaps = 1/110 (0%)
Frame = -1
Query: 673 KIQAHVRGHQVRKNHGKIIWSVGILEKVVLRWRRKGCGLRGFKSEG-VSEGNMVQGESST 497
KIQAHVRGHQVR + IIWSVGILEKV+LRWRRKG GLRGFK E ++EG+ +Q
Sbjct: 524 KIQAHVRGHQVRNKYKNIIWSVGILEKVILRWRRKGSGLRGFKPEATLTEGSNMQDRPVQ 583
Query: 496 EDEYDLLKEGRKQTEQRLQIALARVKSMVQYPEARDQYHRLLNVVTEIQE 347
ED+YD LKEGRKQTEQRLQ ALARVKSMVQYPEARDQY RLLNVV+++++
Sbjct: 584 EDDYDFLKEGRKQTEQRLQKALARVKSMVQYPEARDQYRRLLNVVSDMKD 633
>pir||A84611 hypothetical protein At2g22300 [imported] - Arabidopsis thaliana
gi|4567197|gb|AAD23613.1| unknown protein [Arabidopsis
thaliana]
Length = 1042
Score = 159 bits (402), Expect = 3e-38
Identities = 97/195 (49%), Positives = 116/195 (58%), Gaps = 2/195 (1%)
Frame = -1
Query: 673 KIQAHVRGHQVRKNHGKIIWSVGILEKVVLRWRRKGCGLRGFKSEGVSEGNMVQGESSTE 494
KIQAHVRG+Q RKN+ KIIWSVG+LEKV+LRWRRKG GLRGFKSE + E M G E
Sbjct: 891 KIQAHVRGYQFRKNYRKIIWSVGVLEKVILRWRRKGAGLRGFKSEALVE-KMQDGTEKEE 949
Query: 493 DEYDLLKEGRKQTEQRLQIALARVKSMVQYPEARDQYHRLLNVVTEIQENQVCYPLLFKF 314
D+ D K+GRKQTE RLQ ALARVKSMVQYPEARDQY RLLNVV +IQE++V
Sbjct: 950 DD-DFFKQGRKQTEDRLQKALARVKSMVQYPEARDQYRRLLNVVNDIQESKV-------- 1000
Query: 313 TCCERKIISLKNASRAIVFVIDHRGCLVFFICLLQVKHERTSNDSEET--REFDNLIDLE 140
E+ +SE T + D+LID+E
Sbjct: 1001 --------------------------------------EKALENSEATCFDDDDDLIDIE 1022
Query: 139 ALLDEDIFIQLQLRS 95
ALL++D + L + S
Sbjct: 1023 ALLEDDDTLMLPMSS 1037
>ref|NP_179817.2| ethylene-induced calmodulin-binding protein, putative; protein id:
At2g22300.1 [Arabidopsis thaliana]
Length = 974
Score = 159 bits (402), Expect = 3e-38
Identities = 97/195 (49%), Positives = 116/195 (58%), Gaps = 2/195 (1%)
Frame = -1
Query: 673 KIQAHVRGHQVRKNHGKIIWSVGILEKVVLRWRRKGCGLRGFKSEGVSEGNMVQGESSTE 494
KIQAHVRG+Q RKN+ KIIWSVG+LEKV+LRWRRKG GLRGFKSE + E M G E
Sbjct: 823 KIQAHVRGYQFRKNYRKIIWSVGVLEKVILRWRRKGAGLRGFKSEALVE-KMQDGTEKEE 881
Query: 493 DEYDLLKEGRKQTEQRLQIALARVKSMVQYPEARDQYHRLLNVVTEIQENQVCYPLLFKF 314
D+ D K+GRKQTE RLQ ALARVKSMVQYPEARDQY RLLNVV +IQE++V
Sbjct: 882 DD-DFFKQGRKQTEDRLQKALARVKSMVQYPEARDQYRRLLNVVNDIQESKV-------- 932
Query: 313 TCCERKIISLKNASRAIVFVIDHRGCLVFFICLLQVKHERTSNDSEET--REFDNLIDLE 140
E+ +SE T + D+LID+E
Sbjct: 933 --------------------------------------EKALENSEATCFDDDDDLIDIE 954
Query: 139 ALLDEDIFIQLQLRS 95
ALL++D + L + S
Sbjct: 955 ALLEDDDTLMLPMSS 969
>gb|AAN74651.1| calmodulin-binding transcription factor SR1 [Arabidopsis thaliana]
gi|27311707|gb|AAO00819.1| Unknown protein [Arabidopsis
thaliana]
Length = 1032
Score = 159 bits (402), Expect = 3e-38
Identities = 97/195 (49%), Positives = 116/195 (58%), Gaps = 2/195 (1%)
Frame = -1
Query: 673 KIQAHVRGHQVRKNHGKIIWSVGILEKVVLRWRRKGCGLRGFKSEGVSEGNMVQGESSTE 494
KIQAHVRG+Q RKN+ KIIWSVG+LEKV+LRWRRKG GLRGFKSE + E M G E
Sbjct: 881 KIQAHVRGYQFRKNYRKIIWSVGVLEKVILRWRRKGAGLRGFKSEALVE-KMQDGTEKEE 939
Query: 493 DEYDLLKEGRKQTEQRLQIALARVKSMVQYPEARDQYHRLLNVVTEIQENQVCYPLLFKF 314
D+ D K+GRKQTE RLQ ALARVKSMVQYPEARDQY RLLNVV +IQE++V
Sbjct: 940 DD-DFFKQGRKQTEDRLQKALARVKSMVQYPEARDQYRRLLNVVNDIQESKV-------- 990
Query: 313 TCCERKIISLKNASRAIVFVIDHRGCLVFFICLLQVKHERTSNDSEET--REFDNLIDLE 140
E+ +SE T + D+LID+E
Sbjct: 991 --------------------------------------EKALENSEATCFDDDDDLIDIE 1012
Query: 139 ALLDEDIFIQLQLRS 95
ALL++D + L + S
Sbjct: 1013 ALLEDDDTLMLPMSS 1027
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 556,020,657
Number of Sequences: 1393205
Number of extensions: 11712542
Number of successful extensions: 33989
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 32814
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33940
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29421376608
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)