Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000370A_C01 KMC000370A_c01
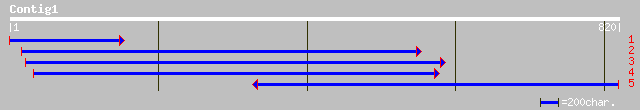
(801 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|Q945F4|IF52_MEDSA Eukaryotic translation initiation factor 5A... 280 2e-74
gb|AAC67555.1| translation initiation factor 5A [Oryza sativa] g... 277 1e-73
sp|Q9SC12|IF5A_SENVE Eukaryotic translation initiation factor 5A... 275 6e-73
gb|AAF27938.1|AF225297_1 translation initiation factor 5A [Eupho... 274 1e-72
sp|P24922|IF52_NICPL Eukaryotic translation initiation factor 5A... 274 1e-72
>sp|Q945F4|IF52_MEDSA Eukaryotic translation initiation factor 5A-2 (eIF-5A 2)
gi|15866587|gb|AAL10404.1|AF416338_1 eukaryotic
translation initiation factor 5A-2 [Medicago sativa]
Length = 159
Score = 280 bits (715), Expect = 2e-74
Identities = 134/144 (93%), Positives = 140/144 (97%)
Frame = -1
Query: 756 ASKTYPQQAGTVRKNGYIVIKNRPCKVVEVSTSKTGKHGHAKCHFVAIDIFTGKKLEDIV 577
ASKTYPQQAGT+RKNGYIVIK+RPCKVVEVSTSKTGKHGHAKCHFVAIDIF GKKLEDIV
Sbjct: 16 ASKTYPQQAGTIRKNGYIVIKSRPCKVVEVSTSKTGKHGHAKCHFVAIDIFNGKKLEDIV 75
Query: 576 PSSHNCDIPHVNRTDYQLIDISEDGFVSLLTDSGDTKDDLKLPTDDALLKQIKEGFGDGK 397
PSSHNCD+PHVNRTDYQLIDISEDGFVSLLTD+G TKDDLKLPTDD+LL QIK+GF DGK
Sbjct: 76 PSSHNCDVPHVNRTDYQLIDISEDGFVSLLTDNGSTKDDLKLPTDDSLLTQIKDGFADGK 135
Query: 396 DLVVSVMSAMGEEQICALKDIGPK 325
DLVVSVMSAMGEEQICALKDIGPK
Sbjct: 136 DLVVSVMSAMGEEQICALKDIGPK 159
>gb|AAC67555.1| translation initiation factor 5A [Oryza sativa]
gi|27817940|dbj|BAC55704.1| translation initiation
factor 5A [Oryza sativa (japonica cultivar-group)]
Length = 160
Score = 277 bits (708), Expect = 1e-73
Identities = 132/144 (91%), Positives = 141/144 (97%)
Frame = -1
Query: 756 ASKTYPQQAGTVRKNGYIVIKNRPCKVVEVSTSKTGKHGHAKCHFVAIDIFTGKKLEDIV 577
ASKTYPQQAGT+RKNGYIVIKNRPCKVVEVSTSKTGKHGHAKCHFVAIDIF GKKLEDIV
Sbjct: 17 ASKTYPQQAGTIRKNGYIVIKNRPCKVVEVSTSKTGKHGHAKCHFVAIDIFNGKKLEDIV 76
Query: 576 PSSHNCDIPHVNRTDYQLIDISEDGFVSLLTDSGDTKDDLKLPTDDALLKQIKEGFGDGK 397
PSSHNCD+PHVNRT+YQLIDISEDGFVSLLT+SG+TKDDL+LPTDD+LL QIK GFG+GK
Sbjct: 77 PSSHNCDVPHVNRTEYQLIDISEDGFVSLLTESGNTKDDLRLPTDDSLLGQIKTGFGEGK 136
Query: 396 DLVVSVMSAMGEEQICALKDIGPK 325
DLVV+VMSAMGEEQICALKDIGPK
Sbjct: 137 DLVVTVMSAMGEEQICALKDIGPK 160
>sp|Q9SC12|IF5A_SENVE Eukaryotic translation initiation factor 5A (eIF-5A)
gi|6689385|emb|CAB65463.1| translation initiation factor
5A precursor protein (eIF-5A) [Senecio vernalis]
Length = 159
Score = 275 bits (703), Expect = 6e-73
Identities = 132/144 (91%), Positives = 140/144 (96%)
Frame = -1
Query: 756 ASKTYPQQAGTVRKNGYIVIKNRPCKVVEVSTSKTGKHGHAKCHFVAIDIFTGKKLEDIV 577
ASKTYPQQAGT+RK G+IVIKNR CKVVEVSTSKTGKHGHAKCHFVAIDIF GKKLEDIV
Sbjct: 16 ASKTYPQQAGTIRKGGHIVIKNRACKVVEVSTSKTGKHGHAKCHFVAIDIFNGKKLEDIV 75
Query: 576 PSSHNCDIPHVNRTDYQLIDISEDGFVSLLTDSGDTKDDLKLPTDDALLKQIKEGFGDGK 397
PSSHNCD+PHVNRTDYQLIDISEDGFVSLLT++G+TKDDLKLPTDDALL QIK+GFG+GK
Sbjct: 76 PSSHNCDVPHVNRTDYQLIDISEDGFVSLLTENGNTKDDLKLPTDDALLTQIKDGFGEGK 135
Query: 396 DLVVSVMSAMGEEQICALKDIGPK 325
DLVVSVMSAMGEEQICALKDIGPK
Sbjct: 136 DLVVSVMSAMGEEQICALKDIGPK 159
>gb|AAF27938.1|AF225297_1 translation initiation factor 5A [Euphorbia esula]
Length = 156
Score = 274 bits (701), Expect = 1e-72
Identities = 129/144 (89%), Positives = 140/144 (96%)
Frame = -1
Query: 756 ASKTYPQQAGTVRKNGYIVIKNRPCKVVEVSTSKTGKHGHAKCHFVAIDIFTGKKLEDIV 577
ASKTYPQQAGT+RKNGYIVIKNRPCKVVEVSTSKTGKHGHAKCHFV IDIF GKKLEDIV
Sbjct: 13 ASKTYPQQAGTIRKNGYIVIKNRPCKVVEVSTSKTGKHGHAKCHFVGIDIFNGKKLEDIV 72
Query: 576 PSSHNCDIPHVNRTDYQLIDISEDGFVSLLTDSGDTKDDLKLPTDDALLKQIKEGFGDGK 397
PSSHNCD+PHV RTDYQLIDISEDGFVSLLT++G+TKDDL+LPTD++LL QIK+GFGDGK
Sbjct: 73 PSSHNCDVPHVTRTDYQLIDISEDGFVSLLTENGNTKDDLRLPTDESLLSQIKDGFGDGK 132
Query: 396 DLVVSVMSAMGEEQICALKDIGPK 325
DLVV+VMS+MGEEQICALKDIGPK
Sbjct: 133 DLVVTVMSSMGEEQICALKDIGPK 156
>sp|P24922|IF52_NICPL Eukaryotic translation initiation factor 5A-2 (eIF-5A) (eIF-4D)
gi|100278|pir||S21059 translation initiation factor
eIF-5A.2 [similarity] - curled-leaved tobacco
gi|19702|emb|CAA45104.1| eukaryotic initiation factor 5A
(2) [Nicotiana plumbaginifolia]
Length = 159
Score = 274 bits (701), Expect = 1e-72
Identities = 131/144 (90%), Positives = 140/144 (96%)
Frame = -1
Query: 756 ASKTYPQQAGTVRKNGYIVIKNRPCKVVEVSTSKTGKHGHAKCHFVAIDIFTGKKLEDIV 577
ASKTYPQQAGT+RKNG+IVIK RPCKVVEVSTSKTGKHGHAKCHFVAIDIFTGKKLEDIV
Sbjct: 16 ASKTYPQQAGTIRKNGHIVIKGRPCKVVEVSTSKTGKHGHAKCHFVAIDIFTGKKLEDIV 75
Query: 576 PSSHNCDIPHVNRTDYQLIDISEDGFVSLLTDSGDTKDDLKLPTDDALLKQIKEGFGDGK 397
PSSHNCD+PHVNRTDYQLIDISEDGFVSLLT++G+TKDDL+LPTDD LL QIK+GF +GK
Sbjct: 76 PSSHNCDVPHVNRTDYQLIDISEDGFVSLLTENGNTKDDLRLPTDDNLLTQIKDGFAEGK 135
Query: 396 DLVVSVMSAMGEEQICALKDIGPK 325
DLVVSVMSAMGEEQICALKDIGPK
Sbjct: 136 DLVVSVMSAMGEEQICALKDIGPK 159
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 656,336,866
Number of Sequences: 1393205
Number of extensions: 14354068
Number of successful extensions: 48188
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 44195
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 47987
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40616159090
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)