Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000358A_C01 KMC000358A_c01
(714 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
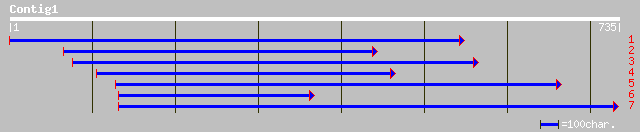
ref|NP_179383.1| hypothetical protein; protein id: At2g17930.1 [... 143 2e-33
ref|NP_680770.1| unknown protein; protein id: At4g36080.1 [Arabi... 141 8e-33
pir||T05501 hypothetical protein T19K4.210 - Arabidopsis thalian... 141 8e-33
pir||C85426 ATM-like protein [imported] - Arabidopsis thaliana g... 141 8e-33
gb|EAA11840.1| ebiP7163 [Anopheles gambiae str. PEST] 42 0.011
>ref|NP_179383.1| hypothetical protein; protein id: At2g17930.1 [Arabidopsis thaliana]
gi|7486992|pir||T00831 hypothetical protein At2g17930
[imported] - Arabidopsis thaliana
Length = 3795
Score = 143 bits (361), Expect = 2e-33
Identities = 63/98 (64%), Positives = 80/98 (81%)
Frame = -2
Query: 713 FFRDELLSWSWRRPLGMPMVPMAAGGTMSPVDFKPKVITNVEHVVARVKGIAPQKFSDEE 534
FFRDELLSW RRPLG+P+ P+ T++P + K KV NVE V+ R++GIAPQ FS+E+
Sbjct: 3698 FFRDELLSWFGRRPLGVPIPPVGGIATLNPAELKHKVNANVEDVIKRIRGIAPQYFSEED 3757
Query: 533 DHVMDPPPSLQRGVTELVEAALNPRNLCMMDPTWHPWF 420
++ ++PP S+QRGV ELVEAAL+PRNLCMMDPTWHPWF
Sbjct: 3758 ENTVEPPQSVQRGVNELVEAALSPRNLCMMDPTWHPWF 3795
>ref|NP_680770.1| unknown protein; protein id: At4g36080.1 [Arabidopsis thaliana]
Length = 3839
Score = 141 bits (356), Expect = 8e-33
Identities = 63/98 (64%), Positives = 82/98 (83%)
Frame = -2
Query: 713 FFRDELLSWSWRRPLGMPMVPMAAGGTMSPVDFKPKVITNVEHVVARVKGIAPQKFSDEE 534
FFRDELLSW RRPLG+P+ P+A T+S + K KV +NV+ V+ R++GIAPQ FS+E+
Sbjct: 3742 FFRDELLSWFGRRPLGVPIPPVAGIATLSSPELKHKVNSNVDDVIGRIRGIAPQYFSEED 3801
Query: 533 DHVMDPPPSLQRGVTELVEAALNPRNLCMMDPTWHPWF 420
++ ++PP S+QRGV+ELVEAAL+PRNLCMMDPTWHPWF
Sbjct: 3802 ENSVEPPQSVQRGVSELVEAALSPRNLCMMDPTWHPWF 3839
>pir||T05501 hypothetical protein T19K4.210 - Arabidopsis thaliana
gi|3036812|emb|CAA18502.1| ATM-like protein [Arabidopsis
thaliana]
Length = 3738
Score = 141 bits (356), Expect = 8e-33
Identities = 63/98 (64%), Positives = 82/98 (83%)
Frame = -2
Query: 713 FFRDELLSWSWRRPLGMPMVPMAAGGTMSPVDFKPKVITNVEHVVARVKGIAPQKFSDEE 534
FFRDELLSW RRPLG+P+ P+A T+S + K KV +NV+ V+ R++GIAPQ FS+E+
Sbjct: 3641 FFRDELLSWFGRRPLGVPIPPVAGIATLSSPELKHKVNSNVDDVIGRIRGIAPQYFSEED 3700
Query: 533 DHVMDPPPSLQRGVTELVEAALNPRNLCMMDPTWHPWF 420
++ ++PP S+QRGV+ELVEAAL+PRNLCMMDPTWHPWF
Sbjct: 3701 ENSVEPPQSVQRGVSELVEAALSPRNLCMMDPTWHPWF 3738
>pir||C85426 ATM-like protein [imported] - Arabidopsis thaliana
gi|7270560|emb|CAB81517.1| ATM-like protein [Arabidopsis
thaliana]
Length = 2089
Score = 141 bits (356), Expect = 8e-33
Identities = 63/98 (64%), Positives = 82/98 (83%)
Frame = -2
Query: 713 FFRDELLSWSWRRPLGMPMVPMAAGGTMSPVDFKPKVITNVEHVVARVKGIAPQKFSDEE 534
FFRDELLSW RRPLG+P+ P+A T+S + K KV +NV+ V+ R++GIAPQ FS+E+
Sbjct: 1992 FFRDELLSWFGRRPLGVPIPPVAGIATLSSPELKHKVNSNVDDVIGRIRGIAPQYFSEED 2051
Query: 533 DHVMDPPPSLQRGVTELVEAALNPRNLCMMDPTWHPWF 420
++ ++PP S+QRGV+ELVEAAL+PRNLCMMDPTWHPWF
Sbjct: 2052 ENSVEPPQSVQRGVSELVEAALSPRNLCMMDPTWHPWF 2089
>gb|EAA11840.1| ebiP7163 [Anopheles gambiae str. PEST]
Length = 3815
Score = 41.6 bits (96), Expect = 0.011
Identities = 14/24 (58%), Positives = 17/24 (70%)
Frame = -2
Query: 494 VTELVEAALNPRNLCMMDPTWHPW 423
++ V A NP NLC+MDP WHPW
Sbjct: 3791 MSTFVHLATNPENLCLMDPAWHPW 3814
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 643,866,629
Number of Sequences: 1393205
Number of extensions: 14312270
Number of successful extensions: 35670
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 34227
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35651
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32936043699
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)