Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000353A_C01 KMC000353A_c01
(633 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
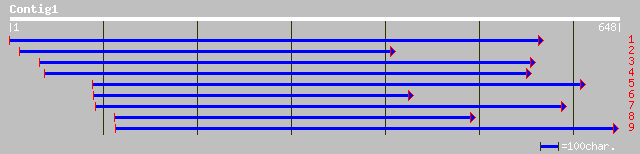
ref|NP_194192.1| putative protein; protein id: At4g24610.1 [Arab... 173 1e-42
ref|NP_201347.2| putative protein; protein id: At5g65440.1, supp... 135 4e-31
dbj|BAB11566.1| gene_id:MNA5.17~pir||T05573~strong similarity to... 135 4e-31
ref|NP_199642.1| putative protein; protein id: At5g48310.1 [Arab... 119 3e-26
gb|ZP_00108451.1| hypothetical protein [Nostoc punctiforme] 34 1.4
>ref|NP_194192.1| putative protein; protein id: At4g24610.1 [Arabidopsis thaliana]
gi|7452447|pir||T05573 hypothetical protein F22K18.190 -
Arabidopsis thaliana gi|4220529|emb|CAA23002.1| putative
protein [Arabidopsis thaliana] gi|7269311|emb|CAB79371.1|
putative protein [Arabidopsis thaliana]
Length = 1145
Score = 173 bits (439), Expect = 1e-42
Identities = 80/102 (78%), Positives = 95/102 (92%)
Frame = -2
Query: 629 DTISHLHSVFETHVFIAICRGYWDRMGQEILSFLENRKENRSWYKGSRVAVSVLDDTFAS 450
+T++HLHSV ETHVFIA+ RGYWDRMGQ +LSFLENRKENR+WYKGSRVAVS+LDDTFA+
Sbjct: 1044 NTVNHLHSVCETHVFIALSRGYWDRMGQIVLSFLENRKENRAWYKGSRVAVSILDDTFAA 1103
Query: 449 QMQQLLGNSLQEKDLDPPRCIMEVRSMLCKDAANHKDNSFYF 324
QMQQLLGNSL+E+DL+PPR IMEVRS+LCKD A++K SFY+
Sbjct: 1104 QMQQLLGNSLREQDLEPPRSIMEVRSILCKDPADNKAKSFYY 1145
>ref|NP_201347.2| putative protein; protein id: At5g65440.1, supported by cDNA:
gi_19715652 [Arabidopsis thaliana]
gi|19715653|gb|AAL91646.1| AT5g65440/MNA5_17 [Arabidopsis
thaliana] gi|22137132|gb|AAM91411.1| At5g65440/MNA5_17
[Arabidopsis thaliana]
Length = 1050
Score = 135 bits (340), Expect = 4e-31
Identities = 59/101 (58%), Positives = 81/101 (79%)
Frame = -2
Query: 626 TISHLHSVFETHVFIAICRGYWDRMGQEILSFLENRKENRSWYKGSRVAVSVLDDTFASQ 447
TI HLH VF VF+AICRG WDRMGQ++L LE+RK+N +W+KG R+AVSVLD+ FA+Q
Sbjct: 950 TIDHLHGVFLPDVFVAICRGIWDRMGQDVLRLLEDRKDNVTWHKGPRIAVSVLDEIFATQ 1009
Query: 446 MQQLLGNSLQEKDLDPPRCIMEVRSMLCKDAANHKDNSFYF 324
MQ LLGN L+ + L+PPR +ME+RSMLCKD+ ++++ + +
Sbjct: 1010 MQSLLGNGLKPEHLEPPRSMMELRSMLCKDSTDYREGGYNY 1050
>dbj|BAB11566.1| gene_id:MNA5.17~pir||T05573~strong similarity to unknown protein
[Arabidopsis thaliana]
Length = 1091
Score = 135 bits (340), Expect = 4e-31
Identities = 59/101 (58%), Positives = 81/101 (79%)
Frame = -2
Query: 626 TISHLHSVFETHVFIAICRGYWDRMGQEILSFLENRKENRSWYKGSRVAVSVLDDTFASQ 447
TI HLH VF VF+AICRG WDRMGQ++L LE+RK+N +W+KG R+AVSVLD+ FA+Q
Sbjct: 991 TIDHLHGVFLPDVFVAICRGIWDRMGQDVLRLLEDRKDNVTWHKGPRIAVSVLDEIFATQ 1050
Query: 446 MQQLLGNSLQEKDLDPPRCIMEVRSMLCKDAANHKDNSFYF 324
MQ LLGN L+ + L+PPR +ME+RSMLCKD+ ++++ + +
Sbjct: 1051 MQSLLGNGLKPEHLEPPRSMMELRSMLCKDSTDYREGGYNY 1091
>ref|NP_199642.1| putative protein; protein id: At5g48310.1 [Arabidopsis thaliana]
gi|8978335|dbj|BAA98188.1|
gene_id:K23F3.3~pir||T05573~strong similarity to unknown
protein [Arabidopsis thaliana]
Length = 1156
Score = 119 bits (299), Expect = 3e-26
Identities = 55/102 (53%), Positives = 78/102 (75%)
Frame = -2
Query: 629 DTISHLHSVFETHVFIAICRGYWDRMGQEILSFLENRKENRSWYKGSRVAVSVLDDTFAS 450
D++S+LH VF + +F+A CR +WDRM Q +L FLE RKEN YKGS A+ +++DTFAS
Sbjct: 1055 DSVSNLHDVFTSQIFVASCRLFWDRMAQVVLKFLEGRKENEVGYKGSYYALGIIEDTFAS 1114
Query: 449 QMQQLLGNSLQEKDLDPPRCIMEVRSMLCKDAANHKDNSFYF 324
+MQ+L GNSLQEKD++ PR ++E RS+L +D N+ ++S YF
Sbjct: 1115 EMQRLQGNSLQEKDMEAPRSVIEARSILSRD--NNANHSSYF 1154
>gb|ZP_00108451.1| hypothetical protein [Nostoc punctiforme]
Length = 409
Score = 34.3 bits (77), Expect = 1.4
Identities = 20/57 (35%), Positives = 31/57 (54%)
Frame = -2
Query: 554 MGQEILSFLENRKENRSWYKGSRVAVSVLDDTFASQMQQLLGNSLQEKDLDPPRCIM 384
+ QE L LENR+ SW K ++ DD+ S+ Q G+ + DLDPP+ ++
Sbjct: 47 LSQEDL-MLENRQAVSSWIKSD---INCGDDSLVSRTLQAKGSKVNRFDLDPPKVLV 99
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 538,087,029
Number of Sequences: 1393205
Number of extensions: 11586369
Number of successful extensions: 25173
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 24507
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25167
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26154777244
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)