Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000349A_C01 KMC000349A_c01
(750 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
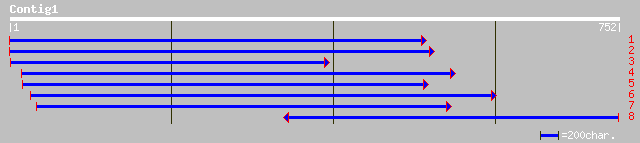
emb|CAB95829.1| hypothetical protein [Cicer arietinum] 200 2e-50
dbj|BAB86220.1| contains ESTs AU093915(E1276),AU162319(E60301)~s... 149 5e-35
ref|NP_173669.1| unknown protein; protein id: At1g22530.1 [Arabi... 134 1e-30
gb|AAK82462.1| At1g72160/T9N14_8 [Arabidopsis thaliana] gi|22137... 134 2e-30
ref|NP_177361.1| cytosolic factor, putative; protein id: At1g721... 134 2e-30
>emb|CAB95829.1| hypothetical protein [Cicer arietinum]
Length = 482
Score = 200 bits (509), Expect = 2e-50
Identities = 96/114 (84%), Positives = 104/114 (91%)
Frame = -2
Query: 749 GEQEFTTADPVTEVTIKPATKHAVELPVSEKGHLAWELRVVGWDVNYGAEFVPGAEDGYT 570
G+QEFTTADP TEVTIKPATKHAVE P+ EK L WE+RVVGWDV+YGAEFVP AEDGYT
Sbjct: 369 GDQEFTTADPATEVTIKPATKHAVEFPIPEKSTLVWEVRVVGWDVSYGAEFVPSAEDGYT 428
Query: 569 VIVQKNRKIGPADETVISNSFKVGEAGKVVLTIDNQTSKKKKLLYRSKTTPISD 408
VIVQKNRKI PADETVI+N+FK+GE GKVVLTIDNQTSKKKKLLYRSKT PIS+
Sbjct: 429 VIVQKNRKIAPADETVINNTFKIGEPGKVVLTIDNQTSKKKKLLYRSKTIPISE 482
>dbj|BAB86220.1| contains ESTs AU093915(E1276),AU162319(E60301)~similar to cytosolic
factor~unknown protein [Oryza sativa (japonica
cultivar-group)] gi|20804751|dbj|BAB92436.1| P0698A10.6
[Oryza sativa (japonica cultivar-group)]
Length = 613
Score = 149 bits (375), Expect = 5e-35
Identities = 70/113 (61%), Positives = 88/113 (76%)
Frame = -2
Query: 746 EQEFTTADPVTEVTIKPATKHAVELPVSEKGHLAWELRVVGWDVNYGAEFVPGAEDGYTV 567
+ EFTT+D VTE+TIKP++K VE+PV+E + WELRV+GW+V+YGAEF P AE GYTV
Sbjct: 499 DPEFTTSDAVTELTIKPSSKETVEIPVTENSTIGWELRVLGWEVSYGAEFTPDAEGGYTV 558
Query: 566 IVQKNRKIGPADETVISNSFKVGEAGKVVLTIDNQTSKKKKLLYRSKTTPISD 408
IVQK RK+ +E ++ SFKVGE GK+VLTI+N SKKKKLLYRSK S+
Sbjct: 559 IVQKTRKVPANEEPIMKGSFKVGEPGKIVLTINNPASKKKKLLYRSKVKSTSE 611
>ref|NP_173669.1| unknown protein; protein id: At1g22530.1 [Arabidopsis thaliana]
gi|25511657|pir||E86358 F12K8.13 protein - Arabidopsis
thaliana gi|6587836|gb|AAF18525.1|AC006551_11 Unknown
protein [Arabidopsis thaliana]
gi|20268782|gb|AAM14094.1| unknown protein [Arabidopsis
thaliana] gi|22136800|gb|AAM91744.1| unknown protein
[Arabidopsis thaliana]
Length = 683
Score = 134 bits (338), Expect = 1e-30
Identities = 65/108 (60%), Positives = 81/108 (74%)
Frame = -2
Query: 746 EQEFTTADPVTEVTIKPATKHAVELPVSEKGHLAWELRVVGWDVNYGAEFVPGAEDGYTV 567
+ FT D VTE +K +K+ ++LP +E L+WELRV+G DV+YGA+F P E YTV
Sbjct: 574 DSPFTVEDGVTEAVVKSTSKYTIDLPATEGSTLSWELRVLGADVSYGAQFEPSNEASYTV 633
Query: 566 IVQKNRKIGPADETVISNSFKVGEAGKVVLTIDNQTSKKKKLLYRSKT 423
IV KNRK+G DE VI++SFK EAGKVV+TIDNQT KKKK+LYRSKT
Sbjct: 634 IVSKNRKVGLTDEPVITDSFKASEAGKVVITIDNQTFKKKKVLYRSKT 681
>gb|AAK82462.1| At1g72160/T9N14_8 [Arabidopsis thaliana] gi|22137076|gb|AAM91383.1|
At1g72160/T9N14_8 [Arabidopsis thaliana]
Length = 390
Score = 134 bits (336), Expect = 2e-30
Identities = 60/109 (55%), Positives = 82/109 (75%)
Frame = -2
Query: 740 EFTTADPVTEVTIKPATKHAVELPVSEKGHLAWELRVVGWDVNYGAEFVPGAEDGYTVIV 561
+F+ D +E+T+KP TK VE+ + EK L WE+RV GW+V+Y AEFVP +D YTV++
Sbjct: 282 DFSLEDSASEITVKPGTKQTVEIIIYEKCELVWEIRVTGWEVSYKAEFVPEEKDAYTVVI 341
Query: 560 QKNRKIGPADETVISNSFKVGEAGKVVLTIDNQTSKKKKLLYRSKTTPI 414
QK RK+ P+DE V+++SFKV E GKV+LT+DN TSKKKKL+YR P+
Sbjct: 342 QKPRKMRPSDEPVLTHSFKVNELGKVLLTVDNPTSKKKKLVYRFNVKPL 390
>ref|NP_177361.1| cytosolic factor, putative; protein id: At1g72160.1, supported by
cDNA: gi_15081613 [Arabidopsis thaliana]
gi|25406150|pir||A96745 probable cytosolic factor
T9N14.8 [imported] - Arabidopsis thaliana
gi|12323663|gb|AAG51796.1|AC067754_12 cytosolic factor,
putative; 19554-17768 [Arabidopsis thaliana]
Length = 490
Score = 134 bits (336), Expect = 2e-30
Identities = 60/109 (55%), Positives = 82/109 (75%)
Frame = -2
Query: 740 EFTTADPVTEVTIKPATKHAVELPVSEKGHLAWELRVVGWDVNYGAEFVPGAEDGYTVIV 561
+F+ D +E+T+KP TK VE+ + EK L WE+RV GW+V+Y AEFVP +D YTV++
Sbjct: 382 DFSLEDSASEITVKPGTKQTVEIIIYEKCELVWEIRVTGWEVSYKAEFVPEEKDAYTVVI 441
Query: 560 QKNRKIGPADETVISNSFKVGEAGKVVLTIDNQTSKKKKLLYRSKTTPI 414
QK RK+ P+DE V+++SFKV E GKV+LT+DN TSKKKKL+YR P+
Sbjct: 442 QKPRKMRPSDEPVLTHSFKVNELGKVLLTVDNPTSKKKKLVYRFNVKPL 490
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 692,009,371
Number of Sequences: 1393205
Number of extensions: 16250438
Number of successful extensions: 53377
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 48470
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 52902
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 36314099463
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)