Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
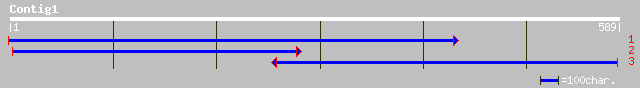
Query= KMC000337A_C01 KMC000337A_c01
(589 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_200643.1| MAP kinase; protein id: At5g58350.1, supported ... 47 2e-04
gb|AAO16559.1| mitogen-activated protein kinase [Triticum aestivum] 44 0.001
gb|AAO18444.1| hypothetical protein [Oryza sativa (japonica cult... 36 0.41
gb|AAM77648.1| toc33 protein [Orychophragmus violaceus] 33 2.7
dbj|BAB85240.1| P0425G02.15 [Oryza sativa (japonica cultivar-gro... 33 3.5
>ref|NP_200643.1| MAP kinase; protein id: At5g58350.1, supported by cDNA:
gi_14532571, supported by cDNA: gi_19548056, supported
by cDNA: gi_20302599 [Arabidopsis thaliana]
gi|8777336|dbj|BAA96926.1| MAP kinase [Arabidopsis
thaliana] gi|14532572|gb|AAK64014.1| unknown protein
[Arabidopsis thaliana] gi|19548057|gb|AAL87392.1|
AT5g58350/AT5g58350 [Arabidopsis thaliana]
gi|20302600|dbj|BAB91127.1| Ser/Thr kinase [Arabidopsis
thaliana]
Length = 571
Score = 47.0 bits (110), Expect = 2e-04
Identities = 41/133 (30%), Positives = 59/133 (43%)
Frame = -2
Query: 585 MNDDESSESSMNSLKCFNFHYCDSGNEEKLNPIFVVGTEHISCSTPKANNKCSGFCPREE 406
++D+ S+SS +S N +Y +P+ + + FCP EE
Sbjct: 442 LHDETYSQSSSHSGSYSNLNYIAVDEYSSQSPVM------------SRTHNMTRFCP-EE 488
Query: 405 DVDVDKPFCNIRIDSHSPRHHGPGPGFRKLTRIPSYVDVRRQQHQPRSLIEEMPQRRMFQ 226
+ N S S R LTR S VDV+RQ RS EE +RR+F+
Sbjct: 489 SSHLQSGQANAYAASSSTNRSLASDN-RTLTRNRSLVDVQRQLLH-RSPGEEARKRRLFK 546
Query: 225 TVGSLENIGFQNP 187
TVG +E +GFQ+P
Sbjct: 547 TVGDVETVGFQSP 559
>gb|AAO16559.1| mitogen-activated protein kinase [Triticum aestivum]
Length = 640
Score = 44.3 bits (103), Expect = 0.001
Identities = 28/69 (40%), Positives = 37/69 (53%), Gaps = 11/69 (15%)
Frame = -2
Query: 354 PRHHGPGPGFR-----------KLTRIPSYVDVRRQQHQPRSLIEEMPQRRMFQTVGSLE 208
PRHH P ++TR S VDVR Q R+L+EE+ +R F TVG++E
Sbjct: 538 PRHHRGSPDAGGDEGRPRRQQGRMTRNRSMVDVRSQLLH-RTLVEELNKRMFFNTVGAVE 596
Query: 207 NIGFQNPEG 181
NIGF+ G
Sbjct: 597 NIGFRRIPG 605
>gb|AAO18444.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 179
Score = 35.8 bits (81), Expect = 0.41
Identities = 17/41 (41%), Positives = 26/41 (62%)
Frame = -2
Query: 285 RQQHQPRSLIEEMPQRRMFQTVGSLENIGFQNPEGGAF*SS 163
R Q R+L+EE+ +R F TVG++ +IGF +P + SS
Sbjct: 5 RSQLLHRTLVEELNKRLFFNTVGAVHDIGFPDPTASSISSS 45
>gb|AAM77648.1| toc33 protein [Orychophragmus violaceus]
Length = 297
Score = 33.1 bits (74), Expect = 2.7
Identities = 25/90 (27%), Positives = 44/90 (48%), Gaps = 13/90 (14%)
Frame = -3
Query: 242 KGGCSRRSVPLRILGFRIPKEVPFEAPGFRNTLISLLLGLYLLPISVNICKPPDLNSSEY 63
KGG + S ++G ++ + PF+A G R ++S +G + ++NI P L + Y
Sbjct: 44 KGGVGKSSTANSLIGEQVVRVSPFQAEGLRPVMVSRTMGGF----TINIIDTPGLVEAGY 99
Query: 62 -----CEIL-------VIMIYVYVKR-DVY 12
E++ I +++YV R DVY
Sbjct: 100 VNHQALELIKGFLVNRTIDVFLYVDRLDVY 129
>dbj|BAB85240.1| P0425G02.15 [Oryza sativa (japonica cultivar-group)]
Length = 354
Score = 32.7 bits (73), Expect = 3.5
Identities = 23/69 (33%), Positives = 30/69 (43%)
Frame = +3
Query: 177 HLLRDSETQYSQGNRPSGTSSFGASPRSEIAVDVVVSSHLHTRVSWSTFEIQVRAHDAGE 356
HLLR+ T+ S RP+ +S GA PRS S H R++W R +G
Sbjct: 7 HLLREFATRNSHPIRPNIRTSSGAGPRSR-------PSGAHARIAW-------RVLASGP 52
Query: 357 SENQFLCCK 383
F C K
Sbjct: 53 PHELFSCAK 61
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 524,116,140
Number of Sequences: 1393205
Number of extensions: 12149798
Number of successful extensions: 35952
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 34836
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35942
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)