Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000332A_C01 KMC000332A_c01
(1070 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
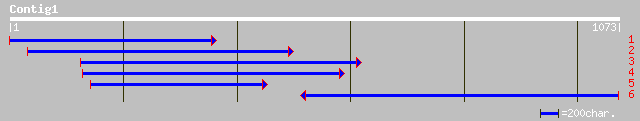
ref|NP_567130.1| putative membrane protein; protein id: At3g6258... 336 3e-91
gb|AAL06485.1|AF411796_1 AT3g62580/T12C14_280 [Arabidopsis thali... 297 2e-79
pir||T48047 hypothetical protein F26K9.10 - Arabidopsis thaliana... 258 1e-67
ref|NP_212133.1| hypothetical protein MBC3205 [Homo sapiens] 101 2e-20
ref|XP_134737.1| hypothetical protein MGC18837 [Mus musculus] gi... 99 1e-19
>ref|NP_567130.1| putative membrane protein; protein id: At3g62580.1, supported by
cDNA: gi_15724186 [Arabidopsis thaliana]
Length = 213
Score = 336 bits (862), Expect = 3e-91
Identities = 161/212 (75%), Positives = 188/212 (87%), Gaps = 5/212 (2%)
Frame = -2
Query: 1066 GWLSRFLTAVTFLAVGVLFSPETLAS-----KSTALSTYLKLAHLLSFSTAFGSALWVTF 902
GW + FL V LAVGV+FSPET S S +S +LKLAHLLSF+TA+G+ALW TF
Sbjct: 2 GWYTSFLVVVASLAVGVIFSPETFGSLTNGENSAKISIFLKLAHLLSFATAWGAALWATF 61
Query: 901 IGGIIMFKNLPRHQFGNLQSKMFPAYFSLVGTCGAIAVASFGYLHPWKTASTSERYQLGF 722
IGGIIMFKNLPRHQFGNLQSK+FPAYF+LVG+C AI++++FGYLHPWK++ST E+YQ+GF
Sbjct: 62 IGGIIMFKNLPRHQFGNLQSKLFPAYFTLVGSCCAISLSAFGYLHPWKSSSTVEKYQIGF 121
Query: 721 LLSSFAFNLTNLFVFTPMTIELMKQRHKVERENNVGEEVGWSKNVEVAKTNPKLAAMNKK 542
LLS+FAFNLTNLFVFTPMTI++MKQRHKVERENN+G+EVGWSKN E AK+ PKLAAMNKK
Sbjct: 122 LLSAFAFNLTNLFVFTPMTIDMMKQRHKVERENNIGDEVGWSKNREKAKSIPKLAAMNKK 181
Query: 541 FGMIHGLSSLSNIMSFGSLAVHSWYLAGKINL 446
FGMIHGLSSL+NI SFGSLA+HSWYLAGK+NL
Sbjct: 182 FGMIHGLSSLANIFSFGSLAMHSWYLAGKLNL 213
>gb|AAL06485.1|AF411796_1 AT3g62580/T12C14_280 [Arabidopsis thaliana]
gi|24111263|gb|AAN46755.1| At3g62580/T12C14_280
[Arabidopsis thaliana]
Length = 206
Score = 297 bits (761), Expect = 2e-79
Identities = 143/190 (75%), Positives = 167/190 (87%), Gaps = 5/190 (2%)
Frame = -2
Query: 1066 GWLSRFLTAVTFLAVGVLFSPETLAS-----KSTALSTYLKLAHLLSFSTAFGSALWVTF 902
GW + FL V LAVGV+FSPET S S +S +LKLAHLLSF+TA+G+ALW TF
Sbjct: 2 GWYTSFLVVVASLAVGVIFSPETFGSLTNGENSAKISIFLKLAHLLSFATAWGAALWATF 61
Query: 901 IGGIIMFKNLPRHQFGNLQSKMFPAYFSLVGTCGAIAVASFGYLHPWKTASTSERYQLGF 722
IGGIIMFKNLPRHQFGNLQSK+FPAYF+LVG+C AI++++FGYLHPWK++ST E+YQ+GF
Sbjct: 62 IGGIIMFKNLPRHQFGNLQSKLFPAYFTLVGSCCAISLSAFGYLHPWKSSSTVEKYQIGF 121
Query: 721 LLSSFAFNLTNLFVFTPMTIELMKQRHKVERENNVGEEVGWSKNVEVAKTNPKLAAMNKK 542
LLS+FAFNLTNLFVFTPMTI++MKQRHKVERENN+G+EVGWSKN E AK+ PKLAAMNKK
Sbjct: 122 LLSAFAFNLTNLFVFTPMTIDMMKQRHKVERENNIGDEVGWSKNREKAKSIPKLAAMNKK 181
Query: 541 FGMIHGLSSL 512
FGMIHGLSSL
Sbjct: 182 FGMIHGLSSL 191
>pir||T48047 hypothetical protein F26K9.10 - Arabidopsis thaliana (fragment)
gi|7362738|emb|CAB83108.1| putative protein [Arabidopsis
thaliana]
Length = 156
Score = 258 bits (658), Expect = 1e-67
Identities = 119/145 (82%), Positives = 139/145 (95%)
Frame = -2
Query: 958 LAHLLSFSTAFGSALWVTFIGGIIMFKNLPRHQFGNLQSKMFPAYFSLVGTCGAIAVASF 779
LAHLLSF+TA+G+ALW TFIGGIIMFKNLPRHQFGNLQSK+FPAYF+LVG+C AI++++F
Sbjct: 1 LAHLLSFATAWGAALWATFIGGIIMFKNLPRHQFGNLQSKLFPAYFTLVGSCCAISLSAF 60
Query: 778 GYLHPWKTASTSERYQLGFLLSSFAFNLTNLFVFTPMTIELMKQRHKVERENNVGEEVGW 599
GYLHPWK++ST E+YQ+GFLLS+FAFNLTNLFVFTPMTI++MKQRHKVERENN+G+EVGW
Sbjct: 61 GYLHPWKSSSTVEKYQIGFLLSAFAFNLTNLFVFTPMTIDMMKQRHKVERENNIGDEVGW 120
Query: 598 SKNVEVAKTNPKLAAMNKKFGMIHG 524
SKN E AK+ PKLAAMNKKFGMIHG
Sbjct: 121 SKNREKAKSIPKLAAMNKKFGMIHG 145
>ref|NP_212133.1| hypothetical protein MBC3205 [Homo sapiens]
Length = 189
Score = 101 bits (252), Expect = 2e-20
Identities = 58/163 (35%), Positives = 82/163 (49%), Gaps = 5/163 (3%)
Frame = -2
Query: 976 LSTYLKLAHLLSFSTAFGSALWVTFIGGIIMFKNLPRHQFGNLQSKMFPAYFSLVGTCGA 797
L +K+ HLL S A+G +WVTF+ G ++F++LPRH FG +QSK+FP YF + C
Sbjct: 7 LGGLIKMVHLLVLSGAWGMQMWVTFVSGFLLFRSLPRHTFGLVQSKLFPFYFHISMGCAF 66
Query: 796 IAVASFGYLHPWKTASTSERYQLGFLLSSFAFNLTNLFVFTPMTIELMKQRHKVERENNV 617
I + H W + E QL L S N P T M VE+E +
Sbjct: 67 INLCILASQHAWAQLTFWEASQLYLLFLSLTLATVNARWLEPRTTAAMWALQTVEKERGL 126
Query: 616 GEEV-----GWSKNVEVAKTNPKLAAMNKKFGMIHGLSSLSNI 503
G EV G ++ + +PK +A+ + F HGLSSL N+
Sbjct: 127 GGEVPGSHQGPDPYRQLREKDPKYSALRQNFFRYHGLSSLCNL 169
>ref|XP_134737.1| hypothetical protein MGC18837 [Mus musculus]
gi|14789776|gb|AAH10787.1| Similar to hypothetical
protein AB030201 [Mus musculus]
Length = 189
Score = 98.6 bits (244), Expect = 1e-19
Identities = 57/159 (35%), Positives = 82/159 (50%), Gaps = 5/159 (3%)
Frame = -2
Query: 964 LKLAHLLSFSTAFGSALWVTFIGGIIMFKNLPRHQFGNLQSKMFPAYFSLVGTCGAIAVA 785
+K+ HLL S A+G +WVTFI G ++F++LPRH FG +QSK+FP YF + C I +
Sbjct: 11 IKVIHLLVLSGAWGMQVWVTFISGFLLFRSLPRHTFGLVQSKVFPVYFHVSLGCAFINLC 70
Query: 784 SFGYLHPWKTASTSERYQLGFLLSSFAFNLTNLFVFTPMTIELMKQRHKVERENNVGEEV 605
W + E QL LL S N T +M+ +E+E +G EV
Sbjct: 71 ILAPQRAWIHLTLWEVSQLSLLLLSLTLATINARWLEARTTAVMRALQSIEKERGLGTEV 130
Query: 604 -----GWSKNVEVAKTNPKLAAMNKKFGMIHGLSSLSNI 503
G ++ +PK +A+ +KF HGLSSL N+
Sbjct: 131 PGNFQGPDPYRQLRDKDPKYSALRRKFYHYHGLSSLCNL 169
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 926,125,047
Number of Sequences: 1393205
Number of extensions: 20747316
Number of successful extensions: 62824
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 58858
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 62754
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 64016183864
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)