Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000326A_C01 KMC000326A_c01
(569 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
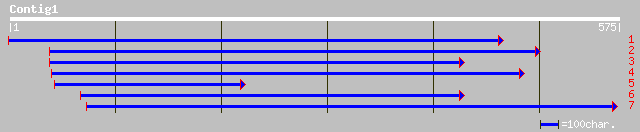
ref|NP_192129.1| drought-induced-19-like 1; protein id: At4g0220... 29 0.32
ref|NP_171775.1| hypothetical protein; protein id: At1g02750.1 [... 34 1.5
dbj|BAC43381.1| unknown protein [Arabidopsis thaliana] 34 1.5
gb|AAL67123.1| AT3g06760/F3E22_10 [Arabidopsis thaliana] gi|2213... 32 7.3
ref|NP_187332.1| unknown protein; protein id: At3g06760.1 [Arabi... 32 7.3
>ref|NP_192129.1| drought-induced-19-like 1; protein id: At4g02200.1, supported by
cDNA: gi_13272462 [Arabidopsis thaliana]
gi|7484943|pir||T01522 drought-induced protein 19
homolog T10M13.20 - Arabidopsis thaliana
gi|3912926|gb|AAC78710.1| drought-induced-19-like 1
[Arabidopsis thaliana] gi|7268980|emb|CAB80713.1|
drought-induced-19-like 1 [Arabidopsis thaliana]
gi|13272463|gb|AAK17170.1|AF325102_1
drought-induced-19-like 1 [Arabidopsis thaliana]
gi|26452996|dbj|BAC43574.1| putative
drought-induced-19-like 1 [Arabidopsis thaliana]
Length = 214
Score = 29.3 bits (64), Expect(2) = 0.32
Identities = 12/26 (46%), Positives = 18/26 (69%)
Frame = -3
Query: 477 LPDQDRIEKARRSKFVQGLLMSTFLD 400
L D + +EKA++ +FVQGL+ S D
Sbjct: 184 LSDTELLEKAKKREFVQGLISSAIFD 209
Score = 25.8 bits (55), Expect(2) = 0.32
Identities = 13/24 (54%), Positives = 15/24 (62%)
Frame = -1
Query: 560 VQPDSSSELSIEDTHSDDTVLERD 489
VQPDSSSE S+ED E+D
Sbjct: 154 VQPDSSSEASMEDNSLIRDSTEKD 177
>ref|NP_171775.1| hypothetical protein; protein id: At1g02750.1 [Arabidopsis
thaliana]
Length = 192
Score = 33.9 bits (76), Expect = 1.5
Identities = 14/28 (50%), Positives = 20/28 (71%)
Frame = -3
Query: 477 LPDQDRIEKARRSKFVQGLLMSTFLDPD 394
L D D++EKA++ +FVQGLL S D +
Sbjct: 160 LSDSDKLEKAKKCEFVQGLLSSAMFDDE 187
>dbj|BAC43381.1| unknown protein [Arabidopsis thaliana]
Length = 221
Score = 33.9 bits (76), Expect = 1.5
Identities = 14/28 (50%), Positives = 20/28 (71%)
Frame = -3
Query: 477 LPDQDRIEKARRSKFVQGLLMSTFLDPD 394
L D D++EKA++ +FVQGLL S D +
Sbjct: 189 LSDSDKLEKAKKCEFVQGLLSSAMFDDE 216
>gb|AAL67123.1| AT3g06760/F3E22_10 [Arabidopsis thaliana]
gi|22137252|gb|AAM91471.1| AT3g06760/F3E22_10
[Arabidopsis thaliana]
Length = 224
Score = 31.6 bits (70), Expect = 7.3
Identities = 13/21 (61%), Positives = 19/21 (89%)
Frame = -3
Query: 456 EKARRSKFVQGLLMSTFLDPD 394
EKA++S+FV+GLL+ST L+ D
Sbjct: 203 EKAKKSEFVRGLLLSTMLEDD 223
>ref|NP_187332.1| unknown protein; protein id: At3g06760.1 [Arabidopsis thaliana]
gi|7549637|gb|AAF63822.1| unknown protein [Arabidopsis
thaliana]
Length = 217
Score = 31.6 bits (70), Expect = 7.3
Identities = 13/21 (61%), Positives = 19/21 (89%)
Frame = -3
Query: 456 EKARRSKFVQGLLMSTFLDPD 394
EKA++S+FV+GLL+ST L+ D
Sbjct: 196 EKAKKSEFVRGLLLSTMLEDD 216
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 431,069,584
Number of Sequences: 1393205
Number of extensions: 8531000
Number of successful extensions: 17508
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 17111
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 17469
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20956655091
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)