Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000320A_C01 KMC000320A_c01
(721 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
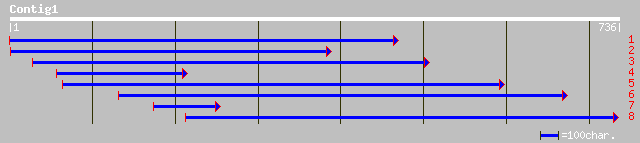
ref|NP_197741.1| putative protein; protein id: At5g23500.1 [Arab... 98 1e-19
dbj|BAB16924.1| P0005A05.28 [Oryza sativa (japonica cultivar-gro... 84 2e-15
emb|CAC08344.1| putative protein [Arabidopsis thaliana] 75 7e-13
ref|NP_197742.1| putative protein; protein id: At5g23510.1 [Arab... 72 8e-12
gb|AAK50128.1|AC087797_13 unknown protein [Oryza sativa] 47 2e-04
>ref|NP_197741.1| putative protein; protein id: At5g23500.1 [Arabidopsis thaliana]
gi|8809710|dbj|BAA97251.1|
gb|AAF01580.1~gene_id:MQM1.24~similar to unknown protein
[Arabidopsis thaliana]
Length = 356
Score = 97.8 bits (242), Expect = 1e-19
Identities = 52/89 (58%), Positives = 66/89 (73%), Gaps = 1/89 (1%)
Frame = -1
Query: 721 LSIGYLDIWEPATLAIKKEGYSIQCHGPNGVVITDKFSPSTPVLIPYGHISEF-IIGSTS 545
L++G++DIWE ATL+IK+EGYSI+C + + I +KFS ST V IP+G +E IIGS
Sbjct: 244 LAVGFVDIWEAATLSIKREGYSIKCI--SDLTIAEKFSASTTVTIPFGQPAELVIIGSDG 301
Query: 544 AEHLLRADNHSTDFSGARDAIVLVLRLFI 458
+EH LRADN S D G+RD IVL LRLFI
Sbjct: 302 SEHSLRADNGSPDLIGSRDEIVLTLRLFI 330
>dbj|BAB16924.1| P0005A05.28 [Oryza sativa (japonica cultivar-group)]
gi|17385662|dbj|BAB78615.1| P0482C06.11 [Oryza sativa
(japonica cultivar-group)]
Length = 737
Score = 84.0 bits (206), Expect = 2e-15
Identities = 47/97 (48%), Positives = 64/97 (65%), Gaps = 2/97 (2%)
Frame = -1
Query: 715 IGYLDIWEPATLAIKKEGYSIQCHGPNGVVITDKFSPSTPVLIPYGHISEFIIGST-SAE 539
+ +LD+WEPA LAIK+EGYSI+C G GVV+T+KF +T + IPYG +EF+I S E
Sbjct: 519 VKFLDMWEPAILAIKREGYSIKCTGQRGVVLTEKFQQATSINIPYGRPTEFLITSADGVE 578
Query: 538 HLLRADNHSTDFSGARDAIVLVLRLF-ILKAGEKKRV 431
+ L+ ++ RD IVLVLRLF I+K R+
Sbjct: 579 YNLKPAENAL----PRDTIVLVLRLFRIMKPTRPDRI 611
>emb|CAC08344.1| putative protein [Arabidopsis thaliana]
Length = 704
Score = 75.5 bits (184), Expect = 7e-13
Identities = 43/74 (58%), Positives = 53/74 (71%), Gaps = 2/74 (2%)
Frame = -1
Query: 715 IGYLDIWEPATLAIKKEGYSIQCHGPNGVVITDKFSPSTPVLIPYGHISEF-IIGSTSAE 539
IGYLDIWE ATL+IKKEGYSI+ N VIT+KFS ST ++IP+ ++F IIG+ E
Sbjct: 535 IGYLDIWEAATLSIKKEGYSIK--PTNDPVITEKFSSSTNIVIPFDQPADFVIIGTDGEE 592
Query: 538 HLLR-ADNHSTDFS 500
HL R DN +TD S
Sbjct: 593 HLCRVVDNDATDLS 606
>ref|NP_197742.1| putative protein; protein id: At5g23510.1 [Arabidopsis thaliana]
gi|8809709|dbj|BAA97250.1|
gb|AAF01580.1~gene_id:MQM1.23~similar to unknown protein
[Arabidopsis thaliana]
Length = 256
Score = 72.0 bits (175), Expect = 8e-12
Identities = 38/73 (52%), Positives = 52/73 (71%), Gaps = 1/73 (1%)
Frame = -1
Query: 721 LSIGYLDIWEPATLAIKKEGYSIQCHGPNGVVITDKFSPSTPVLIPYGHISEF-IIGSTS 545
L+IG++ IWE ATL+I++EGY+I+C+ N + IT+KFS ST V IP+ +E IIGS
Sbjct: 162 LAIGFVHIWEAATLSIEREGYTIKCN--NDLTITEKFSASTAVKIPFEKPAELVIIGSDG 219
Query: 544 AEHLLRADNHSTD 506
+EH LR DN D
Sbjct: 220 SEHCLRVDNEWPD 232
>gb|AAK50128.1|AC087797_13 unknown protein [Oryza sativa]
Length = 505
Score = 47.4 bits (111), Expect = 2e-04
Identities = 35/106 (33%), Positives = 54/106 (50%), Gaps = 7/106 (6%)
Frame = -1
Query: 721 LSIGYL-DIWEPATLAIKKEGYSIQCHGPNGVVITDKFSPSTPVLIPYGHISEFIIGSTS 545
L +GY D WE A L +K+ GY I+ + V+ +K+S + IP G ++F++ S+
Sbjct: 401 LVLGYSSDEWELAILTLKRTGYHIKVK--DEVLTEEKYSSNLQTKIPNGRTTQFVLVSSG 458
Query: 544 AEHL------LRADNHSTDFSGARDAIVLVLRLFILKAGEKKRVKK 425
++ + N+ RD IVLVLR F KA + KR K
Sbjct: 459 GVNIPFNTQGISEPNNEDSDVRLRDLIVLVLRTFQSKALDAKRKGK 504
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 642,469,376
Number of Sequences: 1393205
Number of extensions: 14359975
Number of successful extensions: 36694
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 35245
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36669
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33499052993
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)