Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000316A_C03 KMC000316A_c03
(672 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
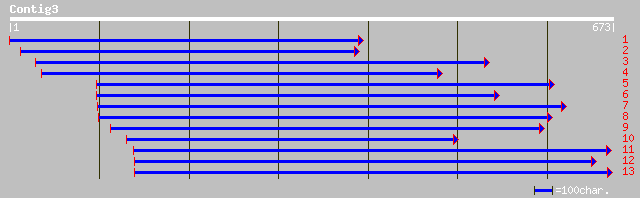
pir||T52306 methionine S-methyltransferase (EC 2.1.1.12) [valida... 139 3e-32
ref|NP_199792.1| methionine S-methyltransferase (gb|AAD49574.1);... 138 6e-32
gb|AAD49573.1| methionine S-methyltransferase [Wollastonia biflora] 127 1e-28
gb|AAD34585.2| S-adenosyl-L-methionine:L-methionine S-methyltran... 119 5e-26
dbj|BAA94795.1| S-adenosyl-L-methionine: L-methionine S-methyltr... 115 5e-25
>pir||T52306 methionine S-methyltransferase (EC 2.1.1.12) [validated] -
Arabidopsis thaliana gi|5733429|gb|AAD49574.1| methionine
S-methyltransferase [Arabidopsis thaliana]
Length = 1071
Score = 139 bits (351), Expect = 3e-32
Identities = 67/126 (53%), Positives = 94/126 (74%)
Frame = -3
Query: 667 DAIVEHIRTLKSRSKCLKEVLEKSGWEVLESWSGVSVVAQPSAYLNKTVKLKIPPKGEGS 488
DA+ E I+TL+ RSK LKEVL+ SGWEV++ +G+S+VA+P AYLNK VKLK
Sbjct: 954 DAVSETIKTLEGRSKRLKEVLQNSGWEVIQPSAGISMVAKPKAYLNKKVKLKA------- 1006
Query: 487 QGNATKEIKLDDSNIRNAILNATGLCINSGSWTGIPGYCRFNIALEENDFQKALDCIPKF 308
G+ + ++L DSN+R+ L+ TG+C+NSGSWTGIPGYCRF+ ALE+++F KA++ I +F
Sbjct: 1007 -GDGQEIVELTDSNMRDVFLSHTGVCLNSGSWTGIPGYCRFSFALEDSEFDKAIESIAQF 1065
Query: 307 QEVALN 290
+ V N
Sbjct: 1066 KSVLAN 1071
>ref|NP_199792.1| methionine S-methyltransferase (gb|AAD49574.1); protein id:
At5g49810.1, supported by cDNA: gi_20453176, supported by
cDNA: gi_5733428 [Arabidopsis thaliana]
gi|8978257|dbj|BAA98148.1| methionine S-methyltransferase
[Arabidopsis thaliana] gi|20453177|gb|AAM19829.1|
AT5g49810/K21G20_2 [Arabidopsis thaliana]
gi|27363322|gb|AAO11580.1| At5g49810/K21G20_2
[Arabidopsis thaliana]
Length = 1071
Score = 138 bits (348), Expect = 6e-32
Identities = 66/126 (52%), Positives = 94/126 (74%)
Frame = -3
Query: 667 DAIVEHIRTLKSRSKCLKEVLEKSGWEVLESWSGVSVVAQPSAYLNKTVKLKIPPKGEGS 488
DA+ E I+TL+ RS+ LKEVL+ SGWEV++ +G+S+VA+P AYLNK VKLK
Sbjct: 954 DAVSETIKTLEGRSRRLKEVLQNSGWEVIQPSAGISMVAKPKAYLNKKVKLKA------- 1006
Query: 487 QGNATKEIKLDDSNIRNAILNATGLCINSGSWTGIPGYCRFNIALEENDFQKALDCIPKF 308
G+ + ++L DSN+R+ L+ TG+C+NSGSWTGIPGYCRF+ ALE+++F KA++ I +F
Sbjct: 1007 -GDGQEIVELTDSNMRDVFLSHTGVCLNSGSWTGIPGYCRFSFALEDSEFDKAIESIAQF 1065
Query: 307 QEVALN 290
+ V N
Sbjct: 1066 KSVLAN 1071
>gb|AAD49573.1| methionine S-methyltransferase [Wollastonia biflora]
Length = 1088
Score = 127 bits (320), Expect = 1e-28
Identities = 64/116 (55%), Positives = 80/116 (68%)
Frame = -3
Query: 646 RTLKSRSKCLKEVLEKSGWEVLESWSGVSVVAQPSAYLNKTVKLKIPPKGEGSQGNATKE 467
+ L +R K LKE LE GWEV+E+ GVSV+A+PSAYL K +KL+ + +T
Sbjct: 979 KLLATRLKRLKETLENCGWEVIEARGGVSVIAKPSAYLGKNIKLE--------KDGSTWV 1030
Query: 466 IKLDDSNIRNAILNATGLCINSGSWTGIPGYCRFNIALEENDFQKALDCIPKFQEV 299
KLD +NIR A+L ATGLCIN SWTGIP YCRF ALE+ DF +ALDCI KF ++
Sbjct: 1031 TKLDGTNIREAMLRATGLCINGPSWTGIPDYCRFTFALEDGDFDRALDCIVKFNQL 1086
>gb|AAD34585.2| S-adenosyl-L-methionine:L-methionine S-methyltransferase [Zea mays]
Length = 1091
Score = 119 bits (297), Expect = 5e-26
Identities = 62/126 (49%), Positives = 80/126 (63%)
Frame = -3
Query: 670 SDAIVEHIRTLKSRSKCLKEVLEKSGWEVLESWSGVSVVAQPSAYLNKTVKLKIPPKGEG 491
SD IVE LK+R+ L + LE GWE G+S++A+P+AY+ K K G
Sbjct: 970 SDLIVEQKEELKNRANQLIQTLESCGWEAAIGCGGISMLAKPTAYMGKAFK------AAG 1023
Query: 490 SQGNATKEIKLDDSNIRNAILNATGLCINSGSWTGIPGYCRFNIALEENDFQKALDCIPK 311
G +LD SNIR AIL ATGLCINS SWTGIPGYCRF+ ALE +F++A+ CI +
Sbjct: 1024 FDG------ELDASNIREAILRATGLCINSSSWTGIPGYCRFSFALERGEFERAMGCIAR 1077
Query: 310 FQEVAL 293
F+E+ L
Sbjct: 1078 FKELVL 1083
>dbj|BAA94795.1| S-adenosyl-L-methionine: L-methionine S-methyltransferase [Hordeum
vulgare subsp. vulgare]
Length = 1088
Score = 115 bits (288), Expect = 5e-25
Identities = 55/126 (43%), Positives = 85/126 (66%)
Frame = -3
Query: 670 SDAIVEHIRTLKSRSKCLKEVLEKSGWEVLESWSGVSVVAQPSAYLNKTVKLKIPPKGEG 491
SD I+E TLK+R+ L ++LE GW+ + G+S++A+P+AY+ K++K+ +G
Sbjct: 965 SDLILEQKETLKNRADQLIKMLESCGWDAVGCHGGISMLAKPTAYIGKSLKV------DG 1018
Query: 490 SQGNATKEIKLDDSNIRNAILNATGLCINSGSWTGIPGYCRFNIALEENDFQKALDCIPK 311
+G KLD N+R A+L +TGLCI+S WTG+P YCRF+ ALE DF +A++CI +
Sbjct: 1019 FEG------KLDSHNMREALLRSTGLCISSSGWTGVPDYCRFSFALESGDFDRAMECIAR 1072
Query: 310 FQEVAL 293
F+E+ L
Sbjct: 1073 FRELVL 1078
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 580,458,116
Number of Sequences: 1393205
Number of extensions: 12472739
Number of successful extensions: 33452
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 32391
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33445
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29421376608
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)