Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000280A_C01 KMC000280A_c01
(780 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
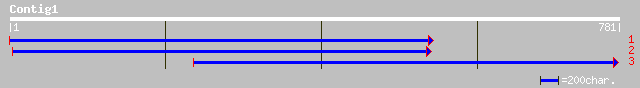
ref|NP_201227.2| calmodulin-binding protein; protein id: At5g642... 256 3e-67
ref|NP_196503.1| calmodulin-binding protein; protein id: At5g094... 245 5e-64
gb|AAM10969.1| calmodulin-binding transcription activator [Brass... 242 5e-63
gb|AAM08530.1|AC079935_2 Putative calmodulin-binding protein sim... 240 2e-62
gb|AAN74651.1| calmodulin-binding transcription factor SR1 [Arab... 221 9e-57
>ref|NP_201227.2| calmodulin-binding protein; protein id: At5g64220.1 [Arabidopsis
thaliana]
Length = 1076
Score = 256 bits (653), Expect = 3e-67
Identities = 134/215 (62%), Positives = 154/215 (71%)
Frame = +2
Query: 74 KKLQFEAQHRWLRPAEICEILRNYRLFQITSEPPNRPPSGSLFLFDRKVLRYFRKDGHSW 253
K+L EAQHRWLRPAEICEILRN++ F I SEPPNRPPSGSLFLFDRKVLRYFRKDGH+W
Sbjct: 22 KQLLSEAQHRWLRPAEICEILRNHQKFHIASEPPNRPPSGSLFLFDRKVLRYFRKDGHNW 81
Query: 254 RKKKDGKTVKEAHEKLKVGSVDALHCYYAHGEESENFQRRSYWMLEQDMMHIVFVHYLDV 433
RKKKDGKTVKEAHEKLKVGS+D LHCYYAHGE++ENFQRR YWMLEQD+MHIVFVHYL+V
Sbjct: 82 RKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGEDNENFQRRCYWMLEQDLMHIVFVHYLEV 141
Query: 434 KVNKTNIGASTDSDGTRSYSHRGRSVSSGFPTNYSSLPSGSTDSMSPTSTLTSLCEDADS 613
K N+ + + GT+ + S+S N S + S S L+ LCEDADS
Sbjct: 142 KGNRMS------TSGTK--ENHSNSLSGTGSVNVDSTATRS-------SILSPLCEDADS 186
Query: 614 EDIHQTSSGLHFFRKSQNMENGQLIDKIDAHSNIS 718
D Q SS L QN E ++ +I H N S
Sbjct: 187 GDSRQASSSL-----QQNPEPQTVVPQIMHHQNAS 216
>ref|NP_196503.1| calmodulin-binding protein; protein id: At5g09410.1 [Arabidopsis
thaliana] gi|9955528|emb|CAC05467.1| putative protein
[Arabidopsis thaliana]
Length = 1007
Score = 245 bits (626), Expect = 5e-64
Identities = 123/190 (64%), Positives = 139/190 (72%)
Frame = +2
Query: 74 KKLQFEAQHRWLRPAEICEILRNYRLFQITSEPPNRPPSGSLFLFDRKVLRYFRKDGHSW 253
++L EAQHRWLRP EICEIL+NY F I SE P RP SGSLFLFDRKVLRYFRKDGH+W
Sbjct: 19 EQLLSEAQHRWLRPTEICEILQNYHKFHIASESPTRPASGSLFLFDRKVLRYFRKDGHNW 78
Query: 254 RKKKDGKTVKEAHEKLKVGSVDALHCYYAHGEESENFQRRSYWMLEQDMMHIVFVHYLDV 433
RKKKDGKT++EAHEKLKVGS+D LHCYYAHGE +ENFQRR YWMLEQ +MHIVFVHYL V
Sbjct: 79 RKKKDGKTIREAHEKLKVGSIDVLHCYYAHGEANENFQRRCYWMLEQHLMHIVFVHYLQV 138
Query: 434 KVNKTNIGASTDSDGTRSYSHRGRSVSSGFPTNYSSLPSGSTDSMSPTSTLTSLCEDADS 613
K N+T+IG + SV+ N S + SPTSTL+SLCEDAD+
Sbjct: 139 KGNRTSIGMK---------ENNSNSVNGTASVNIDS-------TASPTSTLSSLCEDADT 182
Query: 614 EDIHQTSSGL 643
D Q SS L
Sbjct: 183 GDSQQASSVL 192
>gb|AAM10969.1| calmodulin-binding transcription activator [Brassica napus]
Length = 1035
Score = 242 bits (617), Expect = 5e-63
Identities = 122/190 (64%), Positives = 139/190 (72%)
Frame = +2
Query: 74 KKLQFEAQHRWLRPAEICEILRNYRLFQITSEPPNRPPSGSLFLFDRKVLRYFRKDGHSW 253
++L EAQHRWLRPAEICEILRNY F I +E P RP SGSLFLFDRKVL YFRKDGH+W
Sbjct: 19 EQLLSEAQHRWLRPAEICEILRNYHKFHIATESPTRPASGSLFLFDRKVLTYFRKDGHNW 78
Query: 254 RKKKDGKTVKEAHEKLKVGSVDALHCYYAHGEESENFQRRSYWMLEQDMMHIVFVHYLDV 433
RKKKDGKT+KEAHEKLKVGS+D LHCYYAHGE ENFQRR YWMLE ++MHIVFVHYL+V
Sbjct: 79 RKKKDGKTIKEAHEKLKVGSIDVLHCYYAHGEGYENFQRRCYWMLEIELMHIVFVHYLEV 138
Query: 434 KVNKTNIGASTDSDGTRSYSHRGRSVSSGFPTNYSSLPSGSTDSMSPTSTLTSLCEDADS 613
K ++T+IG + S+S N S + SPTS L+S CEDADS
Sbjct: 139 KGSRTSIGMK---------ENNSNSLSGTASVNIDS-------AASPTSRLSSYCEDADS 182
Query: 614 EDIHQTSSGL 643
D HQ+SS L
Sbjct: 183 GDSHQSSSVL 192
>gb|AAM08530.1|AC079935_2 Putative calmodulin-binding protein similar to ER66 [Oryza sativa]
gi|19920231|gb|AAM08663.1|AC113338_19 Putative
calmodulin binding protein similar to ER66 [Oryza sativa
(japonica cultivar-group)]
Length = 1038
Score = 240 bits (612), Expect = 2e-62
Identities = 118/197 (59%), Positives = 140/197 (70%)
Frame = +2
Query: 89 EAQHRWLRPAEICEILRNYRLFQITSEPPNRPPSGSLFLFDRKVLRYFRKDGHSWRKKKD 268
EAQ RWLRPAEICEIL+NY+ F+I EPPNRP SGSLFLFDRKVLRYFRKDGH+WRKKKD
Sbjct: 24 EAQQRWLRPAEICEILKNYKSFRIAPEPPNRPQSGSLFLFDRKVLRYFRKDGHNWRKKKD 83
Query: 269 GKTVKEAHEKLKVGSVDALHCYYAHGEESENFQRRSYWMLEQDMMHIVFVHYLDVKVNKT 448
GKTVKEAHE+LK GS+D LHCYYAHGEE+ENFQRR+YWMLE+D MHIV VHYL+ K K+
Sbjct: 84 GKTVKEAHERLKSGSIDVLHCYYAHGEENENFQRRTYWMLEEDFMHIVLVHYLETKGGKS 143
Query: 449 NIGASTDSDGTRSYSHRGRSVSSGFPTNYSSLPSGSTDSMSPTSTLTSLCEDADSEDIHQ 628
+ D H+ + S S LPS + D S S S E+A+S D++
Sbjct: 144 RTRGNND-------MHQAAVMDSPL----SQLPSQTIDGESSLSGQFSEYEEAESADVYS 192
Query: 629 TSSGLHFFRKSQNMENG 679
+G H F + Q +NG
Sbjct: 193 GGTGYHSFTQMQQQQNG 209
>gb|AAN74651.1| calmodulin-binding transcription factor SR1 [Arabidopsis thaliana]
gi|27311707|gb|AAO00819.1| Unknown protein [Arabidopsis
thaliana]
Length = 1032
Score = 221 bits (563), Expect = 9e-57
Identities = 114/221 (51%), Positives = 146/221 (65%), Gaps = 11/221 (4%)
Frame = +2
Query: 89 EAQHRWLRPAEICEILRNYRLFQITSEPPNRPPSGSLFLFDRKVLRYFRKDGHSWRKKKD 268
EA+HRWLRP EICEIL+NY+ FQI++EPP P SGS+F+FDRKVLRYFRKDGH+WRKKKD
Sbjct: 21 EARHRWLRPPEICEILQNYQRFQISTEPPTTPSSGSVFMFDRKVLRYFRKDGHNWRKKKD 80
Query: 269 GKTVKEAHEKLKVGSVDALHCYYAHGEESENFQRRSYWMLEQDMMHIVFVHYLDVKVNKT 448
GKTVKEAHE+LK GSVD LHCYYAHG+++ENFQRRSYW+L++++ HIVFVHYL+VK ++
Sbjct: 81 GKTVKEAHERLKAGSVDVLHCYYAHGQDNENFQRRSYWLLQEELSHIVFVHYLEVKGSRV 140
Query: 449 NIG---ASTDSDGTRSYSHRGRSVSS--------GFPTNYSSLPSGSTDSMSPTSTLTSL 595
+ D RS G +++S F N S S +TDS S +
Sbjct: 141 STSFNRMQRTEDAARSPQETGDALTSEHDGYASCSFNQNDHSNHSQTTDSASVNGFHSPE 200
Query: 596 CEDADSEDIHQTSSGLHFFRKSQNMENGQLIDKIDAHSNIS 718
EDA+S SS + ++ Q G + D + IS
Sbjct: 201 LEDAESAYNQHGSSTAYSHQELQQPATGGNLTGFDPYYQIS 241
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 672,920,632
Number of Sequences: 1393205
Number of extensions: 14754381
Number of successful extensions: 45240
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 43136
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 45124
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38655378996
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)