Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000246A_C01 KMC000246A_c01
(677 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
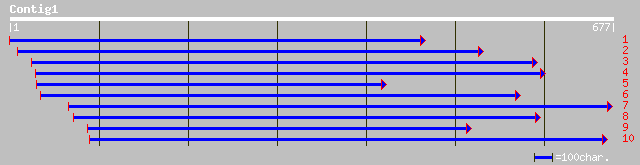
gb|AAL11446.1| HUA enhancer 2 [Arabidopsis thaliana] 89 7e-17
ref|NP_565338.1| DEAD/DEAH box RNA helicase (HUA ENHANCER2); pro... 89 7e-17
pir||B84481 hypothetical protein At2g06990 [imported] - Arabidop... 80 3e-14
ref|NP_056175.1| KIAA0052 protein [Homo sapiens] gi|20306293|gb|... 53 3e-06
gb|AAH14810.1| Similar to RIKEN cDNA 2610528A15 gene [Mus musculus] 53 3e-06
>gb|AAL11446.1| HUA enhancer 2 [Arabidopsis thaliana]
Length = 991
Score = 88.6 bits (218), Expect = 7e-17
Identities = 47/51 (92%), Positives = 47/51 (92%)
Frame = -3
Query: 675 SIIRSARRLDEFLNQLRAAANAVGEVDLEKKFAAASESLRRGIMFANSLYL 523
SIIRSARRLDEFLNQLRAAA AVGE LE KFAAASESLRRGIMFANSLYL
Sbjct: 941 SIIRSARRLDEFLNQLRAAAEAVGESSLESKFAAASESLRRGIMFANSLYL 991
>ref|NP_565338.1| DEAD/DEAH box RNA helicase (HUA ENHANCER2); protein id: At2g06990.1,
supported by cDNA: gi_16024935 [Arabidopsis thaliana]
gi|20197305|gb|AAC67203.2| expressed protein [Arabidopsis
thaliana] gi|28973761|gb|AAO64196.1| putative DEAD/DEAH
box RNA helicase (HUA ENHANCER2) [Arabidopsis thaliana]
Length = 995
Score = 88.6 bits (218), Expect = 7e-17
Identities = 47/51 (92%), Positives = 47/51 (92%)
Frame = -3
Query: 675 SIIRSARRLDEFLNQLRAAANAVGEVDLEKKFAAASESLRRGIMFANSLYL 523
SIIRSARRLDEFLNQLRAAA AVGE LE KFAAASESLRRGIMFANSLYL
Sbjct: 945 SIIRSARRLDEFLNQLRAAAEAVGESSLESKFAAASESLRRGIMFANSLYL 995
>pir||B84481 hypothetical protein At2g06990 [imported] - Arabidopsis thaliana
Length = 996
Score = 79.7 bits (195), Expect = 3e-14
Identities = 47/63 (74%), Positives = 47/63 (74%), Gaps = 12/63 (19%)
Frame = -3
Query: 675 SIIRSARRLDEFLNQ------------LRAAANAVGEVDLEKKFAAASESLRRGIMFANS 532
SIIRSARRLDEFLNQ LRAAA AVGE LE KFAAASESLRRGIMFANS
Sbjct: 934 SIIRSARRLDEFLNQKSDDTLIGDFLQLRAAAEAVGESSLESKFAAASESLRRGIMFANS 993
Query: 531 LYL 523
LYL
Sbjct: 994 LYL 996
>ref|NP_056175.1| KIAA0052 protein [Homo sapiens] gi|20306293|gb|AAH28604.1| KIAA0052
protein [Homo sapiens]
Length = 1042
Score = 53.1 bits (126), Expect = 3e-06
Identities = 27/51 (52%), Positives = 34/51 (65%)
Frame = -3
Query: 675 SIIRSARRLDEFLNQLRAAANAVGEVDLEKKFAAASESLRRGIMFANSLYL 523
SIIR RRL+E L Q+ AA A+G +LE KFA ++R I+FA SLYL
Sbjct: 992 SIIRCMRRLEELLRQMCQAAKAIGNTELENKFAEGITKIKRDIVFAASLYL 1042
>gb|AAH14810.1| Similar to RIKEN cDNA 2610528A15 gene [Mus musculus]
Length = 725
Score = 53.1 bits (126), Expect = 3e-06
Identities = 27/51 (52%), Positives = 34/51 (65%)
Frame = -3
Query: 675 SIIRSARRLDEFLNQLRAAANAVGEVDLEKKFAAASESLRRGIMFANSLYL 523
SIIR RRL+E L Q+ AA A+G +LE KFA ++R I+FA SLYL
Sbjct: 675 SIIRCMRRLEELLRQMCQAAKAIGNTELENKFAEGITKIKRDIVFAASLYL 725
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 575,685,982
Number of Sequences: 1393205
Number of extensions: 12524948
Number of successful extensions: 25938
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 25417
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25930
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29987172312
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)