Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000234A_C01 KMC000234A_c01
(1183 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
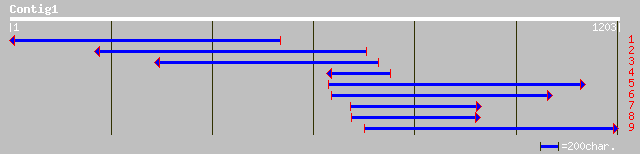
ref|NP_568821.1| putative protein; protein id: At5g55140.1, supp... 139 9e-32
pir||T52585 probable nitrate transporter ntp3 [imported] - Arabi... 119 7e-26
gb|AAM61107.1| nitrate transporter [Arabidopsis thaliana] 119 7e-26
ref|NP_188804.1| nitrate transporter; protein id: At3g21670.1, s... 119 7e-26
emb|CAD41034.1| OSJNBa0060P14.5 [Oryza sativa (japonica cultivar... 97 6e-19
>ref|NP_568821.1| putative protein; protein id: At5g55140.1, supported by cDNA:
92179. [Arabidopsis thaliana] gi|21617966|gb|AAM67016.1|
unknown [Arabidopsis thaliana]
Length = 109
Score = 139 bits (350), Expect = 9e-32
Identities = 63/98 (64%), Positives = 83/98 (84%)
Frame = +3
Query: 90 MNAFKAYQACVPIAWSPNLYITLVRGIPGTRRLHRRTLEALRLGKCNRTVMRWNTPTVRG 269
M+ F+A++A VPI WS +LYITLVRG+PGTR+LHRRTLEA+ L +C+RTV+ N ++RG
Sbjct: 1 MSGFRAFKAQVPIEWSQSLYITLVRGLPGTRKLHRRTLEAMGLRRCHRTVLHSNNSSIRG 60
Query: 270 MLQQVKRLVVIETEEMYKARKQKEEAHRALRPPLVINH 383
M+ QVKR+VV+ETEEMY ARK+ E H+ALRPPLV++H
Sbjct: 61 MIDQVKRMVVVETEEMYNARKEAEANHKALRPPLVVSH 98
>pir||T52585 probable nitrate transporter ntp3 [imported] - Arabidopsis thaliana
(fragment) gi|4490323|emb|CAB38706.1| nitrate transporter
[Arabidopsis thaliana]
Length = 567
Score = 119 bits (299), Expect = 7e-26
Identities = 58/91 (63%), Positives = 73/91 (79%), Gaps = 3/91 (3%)
Frame = -3
Query: 1181 EGMKSMSTGLFLTAISMGYFVSSLLVSIVDKLSKKKWLKSNLNKGRLDYFYWLLAVLGVL 1002
E MKSMSTGLFL+ ISMG+FVSSLLVS+VD+++ K WL+SNLNK RL+YFYWLL VLG L
Sbjct: 461 ERMKSMSTGLFLSTISMGFFVSSLLVSLVDRVTDKSWLRSNLNKARLNYFYWLLVVLGAL 520
Query: 1001 NFILFIVLAMRHHYK---VQHNIEPEDHVDK 918
NF++FIV AM+H YK + + +D V+K
Sbjct: 521 NFLIFIVFAMKHQYKADVITVVVTDDDSVEK 551
>gb|AAM61107.1| nitrate transporter [Arabidopsis thaliana]
Length = 590
Score = 119 bits (299), Expect = 7e-26
Identities = 58/91 (63%), Positives = 73/91 (79%), Gaps = 3/91 (3%)
Frame = -3
Query: 1181 EGMKSMSTGLFLTAISMGYFVSSLLVSIVDKLSKKKWLKSNLNKGRLDYFYWLLAVLGVL 1002
E MKSMSTGLFL+ ISMG+FVSSLLVS+VD+++ K WL+SNLNK RL+YFYWLL VLG L
Sbjct: 484 ERMKSMSTGLFLSTISMGFFVSSLLVSLVDRVTDKSWLRSNLNKARLNYFYWLLVVLGAL 543
Query: 1001 NFILFIVLAMRHHYK---VQHNIEPEDHVDK 918
NF++FIV AM+H YK + + +D V+K
Sbjct: 544 NFLIFIVFAMKHQYKADVITVVVTDDDSVEK 574
>ref|NP_188804.1| nitrate transporter; protein id: At3g21670.1, supported by cDNA:
111089., supported by cDNA: gi_13937208 [Arabidopsis
thaliana] gi|11994403|dbj|BAB02362.1| nitrate transporter
[Arabidopsis thaliana]
gi|13937209|gb|AAK50097.1|AF372959_1 AT3g21670/MIL23_23
[Arabidopsis thaliana] gi|23506051|gb|AAN28885.1|
At3g21670/MIL23_23 [Arabidopsis thaliana]
Length = 590
Score = 119 bits (299), Expect = 7e-26
Identities = 58/91 (63%), Positives = 73/91 (79%), Gaps = 3/91 (3%)
Frame = -3
Query: 1181 EGMKSMSTGLFLTAISMGYFVSSLLVSIVDKLSKKKWLKSNLNKGRLDYFYWLLAVLGVL 1002
E MKSMSTGLFL+ ISMG+FVSSLLVS+VD+++ K WL+SNLNK RL+YFYWLL VLG L
Sbjct: 484 ERMKSMSTGLFLSTISMGFFVSSLLVSLVDRVTDKSWLRSNLNKARLNYFYWLLVVLGAL 543
Query: 1001 NFILFIVLAMRHHYK---VQHNIEPEDHVDK 918
NF++FIV AM+H YK + + +D V+K
Sbjct: 544 NFLIFIVFAMKHQYKADVITVVVTDDDSVEK 574
>emb|CAD41034.1| OSJNBa0060P14.5 [Oryza sativa (japonica cultivar-group)]
Length = 592
Score = 96.7 bits (239), Expect = 6e-19
Identities = 44/75 (58%), Positives = 56/75 (74%)
Frame = -3
Query: 1181 EGMKSMSTGLFLTAISMGYFVSSLLVSIVDKLSKKKWLKSNLNKGRLDYFYWLLAVLGVL 1002
E MKSMSTGLFL ++MG+F+SSLLVS VD ++ W++ L+ GRLD FYW+LA LGV
Sbjct: 489 ERMKSMSTGLFLATLAMGFFLSSLLVSAVDAATRGAWIRDGLDDGRLDLFYWMLAALGVA 548
Query: 1001 NFILFIVLAMRHHYK 957
NF F+V A RH Y+
Sbjct: 549 NFAAFLVFASRHQYR 563
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,000,576,838
Number of Sequences: 1393205
Number of extensions: 22196674
Number of successful extensions: 61852
Number of sequences better than 10.0: 219
Number of HSP's better than 10.0 without gapping: 58377
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 61779
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 73576350696
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)