Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
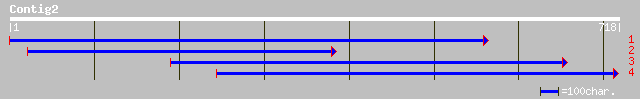
Query= KMC000231A_C02 KMC000231A_c02
(716 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL14248.1| putative heavy-metal transporter [Arabidopsis tha... 40 0.025
ref|NP_179501.1| putative cadmium-transporting ATPase; protein i... 40 0.025
gb|AAL84162.1|AF412407_1 putative heavy metal transporter [Arabi... 40 0.025
ref|NP_194740.1| cadmium-transporting ATPase-like protein; prote... 38 0.16
pir||H71442 probable formyltransferase purU - Arabidopsis thalia... 35 1.4
>gb|AAL14248.1| putative heavy-metal transporter [Arabidopsis thaliana]
Length = 951
Score = 40.4 bits (93), Expect = 0.025
Identities = 26/71 (36%), Positives = 35/71 (48%), Gaps = 5/71 (7%)
Frame = -2
Query: 637 VSACCRNEG----TSKESPESSIVPHACISLDKREVGG-CCKSYMKECCAKHGHGHSGAG 473
V+ CC + TS + S C S ++ G CC+SY KE C+ H H H+ A
Sbjct: 882 VNGCCSSPADLAITSLKVKSDSHCKSNCSSRERCHHGSNCCRSYAKESCS-HDHHHTRAH 940
Query: 472 FVGGLSEIITE 440
VG L EI+ E
Sbjct: 941 GVGTLKEIVIE 951
>ref|NP_179501.1| putative cadmium-transporting ATPase; protein id: At2g19110.1,
supported by cDNA: gi_19071217, supported by cDNA:
gi_20466062 [Arabidopsis thaliana]
gi|12229637|sp|O64474|AHM2_ARATH Potential
cadmium/zinc-transporting ATPase 2
gi|25411904|pir||F84572 probable cadmium-transporting
ATPase [imported] - Arabidopsis thaliana
gi|4210504|gb|AAD12041.1| putative cadmium-transporting
ATPase [Arabidopsis thaliana] gi|11990207|emb|CAC19544.1|
putative heavy metal transporter [Arabidopsis thaliana]
gi|20466063|gb|AAM19954.1| At2g19110/T20K24.12
[Arabidopsis thaliana]
Length = 1172
Score = 40.4 bits (93), Expect = 0.025
Identities = 27/85 (31%), Positives = 37/85 (42%), Gaps = 9/85 (10%)
Frame = -2
Query: 712 CCEDR---SATCHQLLAEFGCEGLSEREVSACCRNEGTSKESPESSI-VPHACISL---- 557
CC D+ + TCH+ A GL ++ C + K + +P AC S
Sbjct: 1078 CCVDKEEVTQTCHEKPASLVVSGLEVKKDEHCESSHRAVKVETCCKVKIPEACASKCRDR 1137
Query: 556 DKREVG-GCCKSYMKECCAKHGHGH 485
KR G CC+SY KE C+ H H
Sbjct: 1138 AKRHSGKSCCRSYAKELCSHRHHHH 1162
>gb|AAL84162.1|AF412407_1 putative heavy metal transporter [Arabidopsis thaliana]
Length = 1172
Score = 40.4 bits (93), Expect = 0.025
Identities = 27/85 (31%), Positives = 37/85 (42%), Gaps = 9/85 (10%)
Frame = -2
Query: 712 CCEDR---SATCHQLLAEFGCEGLSEREVSACCRNEGTSKESPESSI-VPHACISL---- 557
CC D+ + TCH+ A GL ++ C + K + +P AC S
Sbjct: 1078 CCVDKEEVTQTCHEKPASLVVSGLEVKKDEHCESSHRAVKVETCCKVKIPEACASKCRDR 1137
Query: 556 DKREVG-GCCKSYMKECCAKHGHGH 485
KR G CC+SY KE C+ H H
Sbjct: 1138 AKRHSGKSCCRSYAKELCSHRHHHH 1162
>ref|NP_194740.1| cadmium-transporting ATPase-like protein; protein id: At4g30110.1,
supported by cDNA: gi_20384832 [Arabidopsis thaliana]
gi|12229675|sp|Q9SZW4|AHM3_ARATH Potential
cadmium/zinc-transporting ATPase 3 gi|7488066|pir||T08987
probable cadmium-transporting ATPase (EC 3.6.1.-)
F6G3.140 - Arabidopsis thaliana
gi|4938487|emb|CAB43846.1| cadmium-transporting
ATPase-like protein [Arabidopsis thaliana]
gi|7269911|emb|CAB81004.1| cadmium-transporting
ATPase-like protein [Arabidopsis thaliana]
Length = 951
Score = 37.7 bits (86), Expect = 0.16
Identities = 23/58 (39%), Positives = 30/58 (51%), Gaps = 1/58 (1%)
Frame = -2
Query: 610 TSKESPESSIVPHACISLDKREVGG-CCKSYMKECCAKHGHGHSGAGFVGGLSEIITE 440
TS + S C S ++ G CC+SY KE C+ H H H+ A VG L EI+ E
Sbjct: 895 TSLKVKSDSHCKSNCSSRERCHHGSNCCRSYAKESCS-HDHHHTRAHGVGTLKEIVIE 951
>pir||H71442 probable formyltransferase purU - Arabidopsis thaliana
gi|2245095|emb|CAB10517.1| formyltransferase purU
homolog [Arabidopsis thaliana]
gi|7268488|emb|CAB78739.1| formyltransferase purU
homolog [Arabidopsis thaliana]
Length = 295
Score = 34.7 bits (78), Expect = 1.4
Identities = 19/53 (35%), Positives = 29/53 (53%)
Frame = +3
Query: 282 VQTIYKTINSMLPV*ALTVINLKGSFLVIPHHENLLNTNLFKSLAYGGVSYTY 440
V+ ++K + LPV VI+ G F V +HE NT++ + L G+SY Y
Sbjct: 103 VEMLHKWQDGKLPVDITCVISDSGIFGVFSNHERAPNTHVMRFLQRHGISYHY 155
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 630,838,781
Number of Sequences: 1393205
Number of extensions: 14402108
Number of successful extensions: 35809
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 34008
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35728
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33217548346
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)