Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000230A_C01 KMC000230A_c01
(601 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
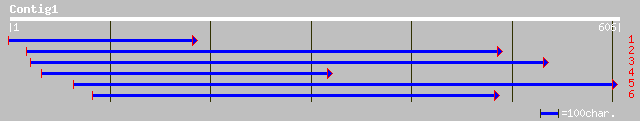
sp|P31384|CCR4_YEAST Glucose-repressible alcohol dehydrogenase t... 33 2.8
gb|AAK06855.1|AF335723_1 hydrogenase-4 component B [Burkholderia... 33 3.7
ref|NP_009381.1| carbon catabolite repression; transcriptional r... 32 4.8
ref|NP_506944.1| Predicted CDS, 7 transmembrane receptor family ... 32 6.2
pir||T18239 transcription effector - yeast (Candida albicans) (f... 32 6.2
>sp|P31384|CCR4_YEAST Glucose-repressible alcohol dehydrogenase transcriptional effector
(Carbon catabolite repressor protein 4)
gi|261567|gb|AAB24455.1| CCR4 [Saccharomyces cerevisiae]
Length = 837
Score = 33.1 bits (74), Expect = 2.8
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Frame = +1
Query: 91 TVKRYHSYCSHWATYNKYNFFPSWFT--PLQRSKAKEPL 201
TV Y++ C H+AT Y + PSW +R+K KE L
Sbjct: 506 TVLSYNTLCQHYATPKMYRYTPSWALSWDYRRNKLKEQL 544
>gb|AAK06855.1|AF335723_1 hydrogenase-4 component B [Burkholderia pseudomallei]
Length = 668
Score = 32.7 bits (73), Expect = 3.7
Identities = 27/78 (34%), Positives = 38/78 (48%), Gaps = 1/78 (1%)
Frame = -3
Query: 383 LFCL-FYIGLLIMSCLYIADCPVFFKLLWWIL*TWGSTFSCLPMLYIE**TRSCEPYFNI 207
L C +++ + M L +AD F + W L T +TF + I R+ YF I
Sbjct: 115 LICFEYHVCVASMVLLLVADDAYCFMVAWETL-TLSATFLVMTNHRIAEIRRAAFVYFLI 173
Query: 206 R*RGSLALLLCKGVNQEG 153
G+LALLLC G+ Q G
Sbjct: 174 SHVGALALLLCFGLLQAG 191
>ref|NP_009381.1| carbon catabolite repression; transcriptional regulator for some
glucose-repressed genes including ADH2; Ccr4p
[Saccharomyces cerevisiae] gi|486964|pir||S36713 CCR4
protein - yeast (Saccharomyces cerevisiae)
gi|171854|gb|AAC04936.1| Ccr4p: Carbon catabolite
repressor protein [Saccharomyces cerevisiae]
Length = 837
Score = 32.3 bits (72), Expect = 4.8
Identities = 15/39 (38%), Positives = 22/39 (55%), Gaps = 2/39 (5%)
Frame = +1
Query: 91 TVKRYHSYCSHWATYNKYNFFPSWFT--PLQRSKAKEPL 201
TV Y++ C H+AT Y + PSW +R+K KE +
Sbjct: 506 TVLSYNTLCQHYATPKMYRYTPSWALSWDYRRNKLKEQI 544
>ref|NP_506944.1| Predicted CDS, 7 transmembrane receptor family member
[Caenorhabditis elegans] gi|7508513|pir||T25302
hypothetical protein T26E4.14 - Caenorhabditis elegans
gi|3880195|emb|CAB03427.1| Hypothetical protein T26E4.14
[Caenorhabditis elegans]
Length = 229
Score = 32.0 bits (71), Expect = 6.2
Identities = 14/41 (34%), Positives = 26/41 (63%)
Frame = -2
Query: 153 EKVVLIICCPM*TIRVISFYCNLLIIVVTPNCYGLKTISTI 31
E +++I+CC T+ V+ NL++++VT C L++I I
Sbjct: 2 ESLLIILCCIYCTVFVVGTTGNLIMVMVTFYCKNLRSICNI 42
>pir||T18239 transcription effector - yeast (Candida albicans) (fragment)
gi|3859723|emb|CAA21997.1| glucose-repressible alcohol
dehydrogenase transcriptional effector [Candida
albicans]
Length = 589
Score = 32.0 bits (71), Expect = 6.2
Identities = 12/24 (50%), Positives = 15/24 (62%)
Frame = +1
Query: 91 TVKRYHSYCSHWATYNKYNFFPSW 162
TV Y++ C H+AT Y F PSW
Sbjct: 259 TVLSYNTLCQHYATPKMYKFTPSW 282
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 515,840,040
Number of Sequences: 1393205
Number of extensions: 11087295
Number of successful extensions: 24001
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 22997
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23993
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)