Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000207A_C01 KMC000207A_c01
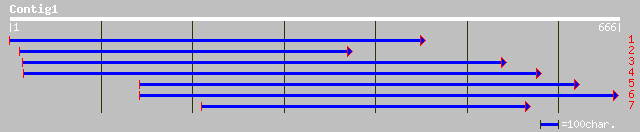
(665 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_172433.1| putative leucyl-tRNA synthetase; protein id: At... 131 7e-30
pir||F96537 hypothetical protein F2J10.2 [imported] - Arabidopsi... 74 1e-12
dbj|BAB14674.1| unnamed protein product [Homo sapiens] 51 1e-05
pir||T51874 hypothetical protein DKFZp762P233.1 - human (fragmen... 51 1e-05
dbj|BAB13817.1| unnamed protein product [Homo sapiens] 51 1e-05
>ref|NP_172433.1| putative leucyl-tRNA synthetase; protein id: At1g09620.1 [Arabidopsis
thaliana] gi|25292955|pir||H86229 hypothetical protein
[imported] - Arabidopsis thaliana
gi|2160156|gb|AAB60719.1| Strong similarity to S. pombe
leucyl-tRNA synthetase (gb|Z73100). [Arabidopsis
thaliana]
Length = 1084
Score = 131 bits (330), Expect = 7e-30
Identities = 67/111 (60%), Positives = 83/111 (74%)
Frame = -3
Query: 663 DSEIMEALQQSSVAPSSNFKQIPKPCMPFLRFKKEEAIKIGAQALDLRLPFGEIEVLREN 484
D+E++ L + + N K I K CMPFL+FKK+EAI IG QAL+LRLPFGEIEVL+ N
Sbjct: 974 DTEMLAELSATLLQEGKNLKAIQKVCMPFLKFKKDEAISIGTQALNLRLPFGEIEVLQSN 1033
Query: 483 LDLIKRQINLEHVEILSAAAADAVAKAGSLASLLNPNPPSPGNPTAIFLTQ 331
DLI+RQ+ LE VEI SA+ D V+ AG ASLL NPPSPG+PTAIF+T+
Sbjct: 1034 KDLIRRQLGLEEVEIYSASDPDDVSIAGPHASLLTQNPPSPGSPTAIFVTR 1084
>pir||F96537 hypothetical protein F2J10.2 [imported] - Arabidopsis thaliana
gi|8569090|gb|AAF76435.1|AC015445_2 Contains similarity
to leucyl tRNA synthetase from Homo sapiens gb|D84223.
[Arabidopsis thaliana]
Length = 843
Score = 74.3 bits (181), Expect = 1e-12
Identities = 34/50 (68%), Positives = 44/50 (88%)
Frame = -3
Query: 585 MPFLRFKKEEAIKIGAQALDLRLPFGEIEVLRENLDLIKRQINLEHVEIL 436
MPF++FKK+EAI IG QAL+LRLPFGEIEVL+ N+DLIKRQ+ LE ++ +
Sbjct: 777 MPFVKFKKKEAISIGIQALNLRLPFGEIEVLKSNMDLIKRQLGLEELKYI 826
>dbj|BAB14674.1| unnamed protein product [Homo sapiens]
Length = 928
Score = 51.2 bits (121), Expect = 1e-05
Identities = 35/116 (30%), Positives = 52/116 (44%), Gaps = 15/116 (12%)
Frame = -3
Query: 633 SSVAPSSNFKQIPKPCMPFLRFKKEEAIKIGAQALDLRLPFGEIEVLRENLDLIKRQINL 454
S + K+ K MPF+ KE K+G + LDL+L F E VL EN+ + + L
Sbjct: 726 SELGSMPELKKYMKKVMPFVAMIKENLEKMGPRILDLQLEFDEKAVLMENIVYLTNSLEL 785
Query: 453 EHVEILSAAAADAVAK---------------AGSLASLLNPNPPSPGNPTAIFLTQ 331
EH+E+ A+ A+ + G SL+NP P + T I + Q
Sbjct: 786 EHIEVKFASEAEDKIREDCCPGKPLNVFRIEPGVSVSLVNPQPSNGHFSTKIEIRQ 841
>pir||T51874 hypothetical protein DKFZp762P233.1 - human (fragment)
gi|9368844|emb|CAB99091.1| hypothetical protein [Homo
sapiens]
Length = 420
Score = 51.2 bits (121), Expect = 1e-05
Identities = 35/116 (30%), Positives = 52/116 (44%), Gaps = 15/116 (12%)
Frame = -3
Query: 633 SSVAPSSNFKQIPKPCMPFLRFKKEEAIKIGAQALDLRLPFGEIEVLRENLDLIKRQINL 454
S + K+ K MPF+ KE K+G + LDL+L F E VL EN+ + + L
Sbjct: 218 SELGSMPELKKYMKKVMPFVAMIKENLEKMGPRILDLQLEFDEKAVLMENIVYLTNSLEL 277
Query: 453 EHVEILSAAAADAVAK---------------AGSLASLLNPNPPSPGNPTAIFLTQ 331
EH+E+ A+ A+ + G SL+NP P + T I + Q
Sbjct: 278 EHIEVKFASEAEDKIREDCCPGKPLNVFRIEPGVSVSLVNPQPSNGHFSTKIEIRQ 333
>dbj|BAB13817.1| unnamed protein product [Homo sapiens]
Length = 438
Score = 51.2 bits (121), Expect = 1e-05
Identities = 35/116 (30%), Positives = 52/116 (44%), Gaps = 15/116 (12%)
Frame = -3
Query: 633 SSVAPSSNFKQIPKPCMPFLRFKKEEAIKIGAQALDLRLPFGEIEVLRENLDLIKRQINL 454
S + K+ K MPF+ KE K+G + LDL+L F E VL EN+ + + L
Sbjct: 236 SELGSMPELKKYMKKVMPFVAMIKENLEKMGPRILDLQLEFDEKAVLMENIVYLTNSLEL 295
Query: 453 EHVEILSAAAADAVAK---------------AGSLASLLNPNPPSPGNPTAIFLTQ 331
EH+E+ A+ A+ + G SL+NP P + T I + Q
Sbjct: 296 EHIEVKFASEAEDKIREDCCPGKPLNVFRIEPGVSVSLVNPQPSNGHFSTKIEIRQ 351
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 539,486,668
Number of Sequences: 1393205
Number of extensions: 11603725
Number of successful extensions: 37227
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 35404
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37155
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28855580904
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)