Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
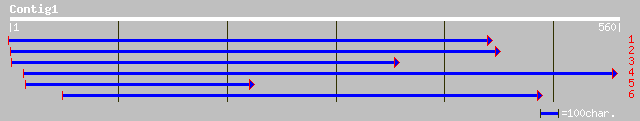
Query= KMC000203A_C01 KMC000203A_c01
(553 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_174005.1| E3 ubiquitin ligase SCF complex subunit Cullin,... 99 8e-46
ref|NP_177125.1| E3 ubiquitin ligase SCF complex subunit Cullin,... 95 8e-45
sp|Q17391|CUL3_CAEEL CUL-3 PROTEIN gi|1381138|gb|AAC47122.1| CUL-3 49 7e-21
ref|NP_503151.1| CULlin CUL-3 (90.2 kD) (cul-3) [Caenorhabditis ... 49 7e-21
ref|NP_523573.1| guftagu CG11861-PA gi|24584423|ref|NP_723907.1|... 60 3e-18
>ref|NP_174005.1| E3 ubiquitin ligase SCF complex subunit Cullin, putative; protein
id: At1g26830.1 [Arabidopsis thaliana]
gi|25354602|pir||A86395 hypothetical protein T2P11.2
[imported] - Arabidopsis thaliana
gi|4262186|gb|AAD14503.1| Highly similar to cullin 3
[Arabidopsis thaliana]
gi|9295728|gb|AAF87034.1|AC006535_12 T24P13.25
[Arabidopsis thaliana]
Length = 732
Score = 98.6 bits (244), Expect(3) = 8e-46
Identities = 46/62 (74%), Positives = 55/62 (88%)
Frame = -3
Query: 551 ALVKGRNVLRKEPMTKDLGEDDAFFVNDKFSSKLYKVKIGTVVAPKESEPEKLETRPRLE 372
A VKG+NV++KEPM+KD+GE+D F VNDKF+SK YKVKIGTVVA KE+EPEK ETR R+E
Sbjct: 601 ACVKGKNVIKKEPMSKDIGEEDLFVVNDKFTSKFYKVKIGTVVAQKETEPEKQETRQRVE 660
Query: 371 ED 366
ED
Sbjct: 661 ED 662
Score = 70.1 bits (170), Expect(3) = 8e-46
Identities = 33/38 (86%), Positives = 35/38 (91%)
Frame = -2
Query: 267 QSRFLANPTEVKKRIESLIEREFLERDDVDRKLYRYLA 154
Q RFLANPTE+KKRIESLIER+FLERD DRKLYRYLA
Sbjct: 695 QPRFLANPTEIKKRIESLIERDFLERDSTDRKLYRYLA 732
Score = 57.8 bits (138), Expect(3) = 8e-46
Identities = 28/32 (87%), Positives = 31/32 (96%)
Frame = -1
Query: 364 RKPQIEAALVRIMKSRKPLDHNHLIAEVTKQL 269
RKPQIEAA+VRIMKSRK LDHN++IAEVTKQL
Sbjct: 663 RKPQIEAAIVRIMKSRKILDHNNIIAEVTKQL 694
>ref|NP_177125.1| E3 ubiquitin ligase SCF complex subunit Cullin, putative; protein
id: At1g69670.1 [Arabidopsis thaliana]
gi|25354600|pir||E96718 probable cullin T6C23.13
[imported] - Arabidopsis thaliana
gi|12325193|gb|AAG52544.1|AC013289_11 putative cullin;
66460-68733 [Arabidopsis thaliana]
Length = 732
Score = 94.7 bits (234), Expect(3) = 8e-45
Identities = 45/62 (72%), Positives = 54/62 (86%)
Frame = -3
Query: 551 ALVKGRNVLRKEPMTKDLGEDDAFFVNDKFSSKLYKVKIGTVVAPKESEPEKLETRPRLE 372
A VKG+NVLRKEPM+K++ E+D F VND+F+SK YKVKIGTVVA KE+EPEK ETR R+E
Sbjct: 601 ACVKGKNVLRKEPMSKEIAEEDWFVVNDRFASKFYKVKIGTVVAQKETEPEKQETRQRVE 660
Query: 371 ED 366
ED
Sbjct: 661 ED 662
Score = 71.2 bits (173), Expect(3) = 8e-45
Identities = 33/38 (86%), Positives = 37/38 (96%)
Frame = -2
Query: 267 QSRFLANPTEVKKRIESLIEREFLERDDVDRKLYRYLA 154
Q+RFLANPTE+KKRIESLIER+FLERD+ DRKLYRYLA
Sbjct: 695 QTRFLANPTEIKKRIESLIERDFLERDNTDRKLYRYLA 732
Score = 57.0 bits (136), Expect(3) = 8e-45
Identities = 27/32 (84%), Positives = 31/32 (96%)
Frame = -1
Query: 364 RKPQIEAALVRIMKSRKPLDHNHLIAEVTKQL 269
RKPQIEAA+VRIMKSR+ LDHN++IAEVTKQL
Sbjct: 663 RKPQIEAAIVRIMKSRRVLDHNNIIAEVTKQL 694
>sp|Q17391|CUL3_CAEEL CUL-3 PROTEIN gi|1381138|gb|AAC47122.1| CUL-3
Length = 780
Score = 48.9 bits (115), Expect(3) = 7e-21
Identities = 22/32 (68%), Positives = 30/32 (93%)
Frame = -1
Query: 364 RKPQIEAALVRIMKSRKPLDHNHLIAEVTKQL 269
RK ++EAA+VRIMK+RK L+HN+L+AEVT+QL
Sbjct: 711 RKLEVEAAIVRIMKARKKLNHNNLVAEVTQQL 742
Score = 47.0 bits (110), Expect(3) = 7e-21
Identities = 25/57 (43%), Positives = 33/57 (57%)
Frame = -3
Query: 536 RNVLRKEPMTKDLGEDDAFFVNDKFSSKLYKVKIGTVVAPKESEPEKLETRPRLEED 366
R ++RK + D F VND F SKL +VK+ V ESEPE ETR ++E+D
Sbjct: 654 RILVRKNKGKDAIDMSDEFAVNDNFQSKLTRVKVQMVTGKVESEPEIRETRQKVEDD 710
Score = 45.8 bits (107), Expect(3) = 7e-21
Identities = 19/36 (52%), Positives = 29/36 (79%)
Frame = -2
Query: 261 RFLANPTEVKKRIESLIEREFLERDDVDRKLYRYLA 154
RF+ +P +K+RIE+LIERE+L RD+ D + Y+Y+A
Sbjct: 745 RFMPSPIIIKQRIETLIEREYLARDEHDHRAYQYIA 780
>ref|NP_503151.1| CULlin CUL-3 (90.2 kD) (cul-3) [Caenorhabditis elegans]
gi|14670190|gb|AAK72067.1|AC024744_1 Hypothetical
protein Y108G3AL.1 [Caenorhabditis elegans]
Length = 777
Score = 48.9 bits (115), Expect(3) = 7e-21
Identities = 22/32 (68%), Positives = 30/32 (93%)
Frame = -1
Query: 364 RKPQIEAALVRIMKSRKPLDHNHLIAEVTKQL 269
RK ++EAA+VRIMK+RK L+HN+L+AEVT+QL
Sbjct: 708 RKLEVEAAIVRIMKARKKLNHNNLVAEVTQQL 739
Score = 47.0 bits (110), Expect(3) = 7e-21
Identities = 25/57 (43%), Positives = 33/57 (57%)
Frame = -3
Query: 536 RNVLRKEPMTKDLGEDDAFFVNDKFSSKLYKVKIGTVVAPKESEPEKLETRPRLEED 366
R ++RK + D F VND F SKL +VK+ V ESEPE ETR ++E+D
Sbjct: 651 RILVRKNKGKDAIDMSDEFAVNDNFQSKLTRVKVQMVTGKVESEPEIRETRQKVEDD 707
Score = 45.8 bits (107), Expect(3) = 7e-21
Identities = 19/36 (52%), Positives = 29/36 (79%)
Frame = -2
Query: 261 RFLANPTEVKKRIESLIEREFLERDDVDRKLYRYLA 154
RF+ +P +K+RIE+LIERE+L RD+ D + Y+Y+A
Sbjct: 742 RFMPSPIIIKQRIETLIEREYLARDEHDHRAYQYIA 777
>ref|NP_523573.1| guftagu CG11861-PA gi|24584423|ref|NP_723907.1| guftagu CG11861-PB
gi|7287895|gb|AAF44933.1|AE003406_138 symbol=gft;
synonym=BG:DS07851.2; cDNA=method:''sim4'',
score:''1000.0'', desc:''LD10516 LD Drosophila
melanogaster embryo BlueScript Drosophila melanogaster
cDNA clone, full length mRNA sequence from BDGP'';
match=method:''BLASTX'', version:''2.0a19MP-WashU
[05-Feb-1998] [Build sol2.5-ultra 01:47:30
05-Feb-1998]'', score:''832.0'',
desc:''trEMBL::d1032553:KIAA0617 PROTEIN. organism:HOMO
SAPIENS (HUMAN). dbxref:GenBank; AB014517; d1032553;
-.'', species:''HOMO SAPIENS gi|7298217|gb|AAF53450.1|
CG11861-PA [Drosophila melanogaster]
gi|7298218|gb|AAF53451.1| CG11861-PB [Drosophila
melanogaster]
Length = 773
Score = 59.7 bits (143), Expect(2) = 3e-18
Identities = 32/73 (43%), Positives = 44/73 (59%)
Frame = -3
Query: 530 VLRKEPMTKDLGEDDAFFVNDKFSSKLYKVKIGTVVAPKESEPEKLETRPRLEEDTEAAD 351
V + TKD+ D F+VND F+SK ++VKI TV A ESEPE+ ETR +++ED +
Sbjct: 649 VRNSKTKTKDIEPTDEFYVNDAFNSKFHRVKIQTVAAKGESEPERKETRGKVDEDRK--- 705
Query: 350 *GSASEDHEVEKA 312
HE+E A
Sbjct: 706 -------HEIEAA 711
Score = 53.5 bits (127), Expect(2) = 3e-18
Identities = 25/41 (60%), Positives = 31/41 (74%)
Frame = -2
Query: 276 SSCQSRFLANPTEVKKRIESLIEREFLERDDVDRKLYRYLA 154
S +SRFL +P +KKRIE LIERE+L+R DRK+Y YLA
Sbjct: 733 SQLKSRFLPSPVFIKKRIEGLIEREYLQRSPEDRKVYNYLA 773
Score = 45.8 bits (107), Expect = 3e-04
Identities = 24/41 (58%), Positives = 32/41 (77%)
Frame = -1
Query: 391 KLDQDWRRTRKPQIEAALVRIMKSRKPLDHNHLIAEVTKQL 269
K+D+D RK +IEAA+VRIMK+RK L HN L+++VT QL
Sbjct: 699 KVDED----RKHEIEAAIVRIMKARKRLAHNLLVSDVTSQL 735
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 453,435,391
Number of Sequences: 1393205
Number of extensions: 9230149
Number of successful extensions: 26340
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 25061
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26266
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19234190289
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)