Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
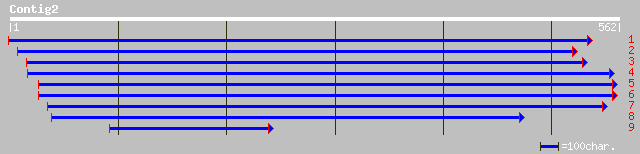
Query= KMC000193A_C02 KMC000193A_c02
(561 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF61649.1|AF200304_2 beta-1,4-xylanase XynA precursor [Caldi... 34 1.1
ref|XP_230116.1| similar to olfactory receptor MOR179-1 [Mus mus... 33 1.8
ref|NP_344144.1| Formate dehydrogenase Alpha subunit (fdhF-2) [S... 33 1.8
ref|NP_567207.1| hypothetical protein; protein id: At4g01023.1 [... 33 3.1
pir||T10542 hypothetical protein F3I3.40 - Arabidopsis thaliana ... 33 3.1
>gb|AAF61649.1|AF200304_2 beta-1,4-xylanase XynA precursor [Caldibacillus cellulovorans]
Length = 921
Score = 34.3 bits (77), Expect = 1.1
Identities = 35/127 (27%), Positives = 52/127 (40%), Gaps = 4/127 (3%)
Frame = +3
Query: 156 RMQQNQKLPKKYSVALPTSKTITSVVQMHERPQSTTSQ--EKGEQIKRKPILKH-N*SKI 326
R++Q LPK +P+ K + + P T + E ++ RK ILKH N
Sbjct: 180 RIEQLPDLPKTVEENIPSLKDVFA----GRFPIGTAFENFELLDEQDRKLILKHFNSVTP 235
Query: 327 IHICKWSTFHSWEGELVKRRHMGSHCVEFAITRA-KTVTLSVQTAMQSVRRNAVWFTKQP 503
++ KW + EG F T + K V +VQ M+ +W + P
Sbjct: 236 GNVLKWDSTEPQEGV-------------FNFTESDKAVAFAVQNGMKIRGHTLIWHNQTP 282
Query: 504 NWVTYIS 524
NWV Y S
Sbjct: 283 NWVFYDS 289
>ref|XP_230116.1| similar to olfactory receptor MOR179-1 [Mus musculus] [Rattus
norvegicus]
Length = 315
Score = 33.5 bits (75), Expect = 1.8
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 3/53 (5%)
Frame = -1
Query: 252 AVFHAFAPLRLSFCSLGELQNIFLGVSG-SAASCQHNYRESML--YLSNFISL 103
A+ H A RLSFC E+++IF + A SC Y +L + +FI L
Sbjct: 154 AIIHTVATFRLSFCQSNEIKHIFCDIPPLLAISCSETYINELLLFFFVSFIEL 206
>ref|NP_344144.1| Formate dehydrogenase Alpha subunit (fdhF-2) [Sulfolobus
solfataricus] gi|25387635|pir||G90459 formate
dehydrogenase Alpha subunit (fdhF-2) [imported] -
Sulfolobus solfataricus gi|13816176|gb|AAK42934.1|
Formate dehydrogenase Alpha subunit (fdhF-2) [Sulfolobus
solfataricus]
Length = 979
Score = 33.5 bits (75), Expect = 1.8
Identities = 21/53 (39%), Positives = 25/53 (46%), Gaps = 2/53 (3%)
Frame = +2
Query: 404 CRICHNPRQNCDLIGA--NCNAVSQEKCSLVYQTTKLGNLYIC*PHGCILLGQ 556
C IC N +C L A N SQ+ YQT + G YI P CIL G+
Sbjct: 100 CSICENNNGDCVLHEAVIKLNINSQKYVEKPYQTDESGPFYIYDPSQCILCGR 152
>ref|NP_567207.1| hypothetical protein; protein id: At4g01023.1 [Arabidopsis
thaliana]
Length = 236
Score = 32.7 bits (73), Expect = 3.1
Identities = 18/43 (41%), Positives = 23/43 (52%), Gaps = 1/43 (2%)
Frame = +2
Query: 374 GEEKTHGIPL-CRICHNPRQNCDLIGANCNAVSQEKCSLVYQT 499
GEE +PL C IC NP D + NCN +KC+L + T
Sbjct: 75 GEEDDDALPLACSICQNPF--LDPVVTNCNHYFCDKCALKHHT 115
>pir||T10542 hypothetical protein F3I3.40 - Arabidopsis thaliana
gi|5262156|emb|CAB45785.1| putative protein [Arabidopsis
thaliana] gi|7267599|emb|CAB80911.1| putative protein
[Arabidopsis thaliana]
Length = 2322
Score = 32.7 bits (73), Expect = 3.1
Identities = 18/43 (41%), Positives = 23/43 (52%), Gaps = 1/43 (2%)
Frame = +2
Query: 374 GEEKTHGIPL-CRICHNPRQNCDLIGANCNAVSQEKCSLVYQT 499
GEE +PL C IC NP D + NCN +KC+L + T
Sbjct: 1907 GEEDDDALPLACSICQNPF--LDPVVTNCNHYFCDKCALKHHT 1947
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 491,908,891
Number of Sequences: 1393205
Number of extensions: 10980201
Number of successful extensions: 26468
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 25749
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26457
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)