Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
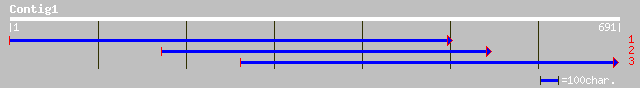
Query= KMC000182A_C01 KMC000182A_c01
(691 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_196024.1| expressed protein; protein id: At5g04040.1 [Ara... 46 4e-06
ref|NP_191273.1| expressed protein; protein id: At3g57140.1 [Ara... 42 0.010
dbj|BAC37140.1| unnamed protein product [Mus musculus] 35 0.75
ref|NP_035012.1| necdin [Mus musculus] gi|1040691|dbj|BAA11183.1... 35 0.75
ref|XP_218733.1| similar to necdin [Mus musculus] [Rattus norveg... 35 0.75
>ref|NP_196024.1| expressed protein; protein id: At5g04040.1 [Arabidopsis thaliana]
gi|11282314|pir||T48431 hypothetical protein F8F6.250 -
Arabidopsis thaliana gi|7406414|emb|CAB85524.1| putative
protein [Arabidopsis thaliana]
gi|22531263|gb|AAM97135.1| putative protein [Arabidopsis
thaliana]
Length = 825
Score = 45.8 bits (107), Expect(2) = 4e-06
Identities = 26/55 (47%), Positives = 32/55 (57%), Gaps = 1/55 (1%)
Frame = -3
Query: 599 NGIVFNVVQKEGL-RPSNRSHDFGNYRNEVAECVPIDCPEKEMDAASSASEPEDD 438
NG V NVV++E L PS GN E+ E V +D PEKEMD +S + EDD
Sbjct: 743 NGFVLNVVKRENLGMPS-----IGNQNTELPESVQLDIPEKEMDCSSVSEHEEDD 792
Score = 26.6 bits (57), Expect(2) = 4e-06
Identities = 12/15 (80%), Positives = 12/15 (80%)
Frame = -2
Query: 651 SSILVTEGDLFPPER 607
SSI VTEGDL PER
Sbjct: 726 SSITVTEGDLLQPER 740
>ref|NP_191273.1| expressed protein; protein id: At3g57140.1 [Arabidopsis thaliana]
gi|11282313|pir||T47774 hypothetical protein F24I3.220 -
Arabidopsis thaliana gi|6911884|emb|CAB72184.1| putative
protein [Arabidopsis thaliana]
gi|26450904|dbj|BAC42559.1| unknown protein [Arabidopsis
thaliana] gi|29029050|gb|AAO64904.1| At3g57140
[Arabidopsis thaliana]
Length = 801
Score = 41.6 bits (96), Expect = 0.010
Identities = 25/61 (40%), Positives = 35/61 (56%), Gaps = 3/61 (4%)
Frame = -3
Query: 605 SHNGIVFNVVQKEGLRPSNRSHDFGNYRN--EVAECVPIDCPEKE-MDAASSASEPEDDE 435
+HNG V N+V+ E LR ++ D N E E V +D PEK+ +D SSASE D +
Sbjct: 734 THNGFVLNLVRGENLRMNSEPEDSQNESEIPETPESVQLDSPEKDIIDGESSASEDGDAQ 793
Query: 434 S 432
+
Sbjct: 794 A 794
>dbj|BAC37140.1| unnamed protein product [Mus musculus]
Length = 325
Score = 35.4 bits (80), Expect = 0.75
Identities = 17/51 (33%), Positives = 30/51 (58%)
Frame = -3
Query: 659 LLGLVFW*LKVIFSRLKGSHNGIVFNVVQKEGLRPSNRSHDFGNYRNEVAE 507
+ GL+ L +I+ + +G+ G V+NV++ GLRP + FG+ R + E
Sbjct: 191 MTGLLLMILSLIYVKGRGAREGAVWNVLRILGLRPWKKHSTFGDVRKIITE 241
>ref|NP_035012.1| necdin [Mus musculus] gi|1040691|dbj|BAA11183.1| necdin [Mus
musculus]
Length = 325
Score = 35.4 bits (80), Expect = 0.75
Identities = 17/51 (33%), Positives = 30/51 (58%)
Frame = -3
Query: 659 LLGLVFW*LKVIFSRLKGSHNGIVFNVVQKEGLRPSNRSHDFGNYRNEVAE 507
+ GL+ L +I+ + +G+ G V+NV++ GLRP + FG+ R + E
Sbjct: 191 MTGLLLMILSLIYVKGRGAREGAVWNVLRILGLRPWKKHSTFGDVRKIITE 241
>ref|XP_218733.1| similar to necdin [Mus musculus] [Rattus norvegicus]
Length = 325
Score = 35.4 bits (80), Expect = 0.75
Identities = 17/51 (33%), Positives = 30/51 (58%)
Frame = -3
Query: 659 LLGLVFW*LKVIFSRLKGSHNGIVFNVVQKEGLRPSNRSHDFGNYRNEVAE 507
+ GL+ L +I+ + +G+ G V+NV++ GLRP + FG+ R + E
Sbjct: 191 MTGLLLMILSLIYVKGRGAREGAVWNVLRILGLRPWKKHSTFGDVRKIITE 241
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 628,096,962
Number of Sequences: 1393205
Number of extensions: 14168375
Number of successful extensions: 33331
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 31853
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33278
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 31118763720
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)