Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000179A_C01 KMC000179A_c01
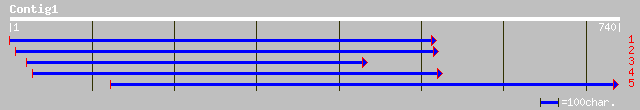
(738 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF79445.1|AC025808_27 F18O14.22 [Arabidopsis thaliana] 225 4e-58
ref|NP_173377.1| integral membrane protein, putative; protein id... 225 4e-58
ref|NP_177658.1| integral membrane protein, putative; protein id... 225 6e-58
pir||T14545 probable sugar transporter protein - beet gi|1209756... 217 2e-55
gb|AAD39600.1|AC007858_14 Similar to gb|U43629 integral membrane... 212 4e-54
>gb|AAF79445.1|AC025808_27 F18O14.22 [Arabidopsis thaliana]
Length = 515
Score = 225 bits (574), Expect = 4e-58
Identities = 107/152 (70%), Positives = 131/152 (85%)
Frame = -2
Query: 737 STWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMV 558
+TWLVDK+GRRLLL++SS MT+SLV+V++AFYL+ VS DS++Y IL +VSVVG+V MV
Sbjct: 364 ATWLVDKAGRRLLLMISSIGMTISLVIVAVAFYLKEFVSPDSNMYNILSMVSVVGVVAMV 423
Query: 557 IGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGGTFTM 378
I SLG+GPIPW+IMSEILPV+IKGLAGS AT+ NW SW +TMTAN+LL WSSGGTFT+
Sbjct: 424 ISCSLGMGPIPWLIMSEILPVNIKGLAGSIATLLNWFVSWLVTMTANMLLAWSSGGTFTL 483
Query: 377 YTVVAAFTVVFTALWVPETKGRTLEEIQSSFR 282
Y +V FTVVF +LWVPETKG+TLEEIQ+ FR
Sbjct: 484 YALVCGFTVVFVSLWVPETKGKTLEEIQALFR 515
>ref|NP_173377.1| integral membrane protein, putative; protein id: At1g19450.1,
supported by cDNA: gi_16648956, supported by cDNA:
gi_20259851 [Arabidopsis thaliana]
gi|16648957|gb|AAL24330.1| similar to integral membrane
protein [Arabidopsis thaliana]
gi|20259852|gb|AAM13273.1| similar to integral membrane
protein [Arabidopsis thaliana]
Length = 488
Score = 225 bits (574), Expect = 4e-58
Identities = 107/152 (70%), Positives = 131/152 (85%)
Frame = -2
Query: 737 STWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMV 558
+TWLVDK+GRRLLL++SS MT+SLV+V++AFYL+ VS DS++Y IL +VSVVG+V MV
Sbjct: 337 ATWLVDKAGRRLLLMISSIGMTISLVIVAVAFYLKEFVSPDSNMYNILSMVSVVGVVAMV 396
Query: 557 IGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGGTFTM 378
I SLG+GPIPW+IMSEILPV+IKGLAGS AT+ NW SW +TMTAN+LL WSSGGTFT+
Sbjct: 397 ISCSLGMGPIPWLIMSEILPVNIKGLAGSIATLLNWFVSWLVTMTANMLLAWSSGGTFTL 456
Query: 377 YTVVAAFTVVFTALWVPETKGRTLEEIQSSFR 282
Y +V FTVVF +LWVPETKG+TLEEIQ+ FR
Sbjct: 457 YALVCGFTVVFVSLWVPETKGKTLEEIQALFR 488
>ref|NP_177658.1| integral membrane protein, putative; protein id: At1g75220.1,
supported by cDNA: gi_15724239 [Arabidopsis thaliana]
gi|25308944|pir||E96782 hypothetical protein F22H5.6
[imported] - Arabidopsis thaliana
gi|10092276|gb|AAG12689.1|AC025814_13 integral membrane
protein, putative; 33518-36712 [Arabidopsis thaliana]
gi|15724240|gb|AAL06513.1|AF412060_1 At1g75220/F22H5_6
[Arabidopsis thaliana] gi|21700861|gb|AAM70554.1|
At1g75220/F22H5_6 [Arabidopsis thaliana]
Length = 487
Score = 225 bits (573), Expect = 6e-58
Identities = 111/152 (73%), Positives = 129/152 (84%)
Frame = -2
Query: 737 STWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMV 558
STWLVDK+GRRLLL +SS MT+SLV+V+ AFYL+ VS DS +Y L I+SVVG+V MV
Sbjct: 336 STWLVDKAGRRLLLTISSVGMTISLVIVAAAFYLKEFVSPDSDMYSWLSILSVVGVVAMV 395
Query: 557 IGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGGTFTM 378
+ FSLG+GPIPW+IMSEILPV+IKGLAGS AT+ANW SW ITMTANLLL WSSGGTFT+
Sbjct: 396 VFFSLGMGPIPWLIMSEILPVNIKGLAGSIATLANWFFSWLITMTANLLLAWSSGGTFTL 455
Query: 377 YTVVAAFTVVFTALWVPETKGRTLEEIQSSFR 282
Y +V AFTVVF LWVPETKG+TLEE+QS FR
Sbjct: 456 YGLVCAFTVVFVTLWVPETKGKTLEELQSLFR 487
>pir||T14545 probable sugar transporter protein - beet gi|1209756|gb|AAB53155.1|
integral membrane protein [Beta vulgaris]
Length = 490
Score = 217 bits (552), Expect = 2e-55
Identities = 101/152 (66%), Positives = 131/152 (85%)
Frame = -2
Query: 737 STWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMV 558
+TWLVDKSGRRLLL++SSS MT+SL+VV+++F+L+ VS++S Y + I+SVVG+V MV
Sbjct: 338 TTWLVDKSGRRLLLIVSSSGMTLSLLVVAMSFFLKEMVSDESTWYSVFSILSVVGVVAMV 397
Query: 557 IGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGGTFTM 378
+ FSLG+G IPW+IMSEILP++IKGLAGS AT+ANW +W +TMTAN++L+W+SGGTF++
Sbjct: 398 VTFSLGIGAIPWIIMSEILPINIKGLAGSIATLANWFVAWIVTMTANIMLSWNSGGTFSI 457
Query: 377 YTVVAAFTVVFTALWVPETKGRTLEEIQSSFR 282
Y VV AFTV F +WVPETKGRTLEEIQ SFR
Sbjct: 458 YMVVCAFTVAFVVIWVPETKGRTLEEIQWSFR 489
>gb|AAD39600.1|AC007858_14 Similar to gb|U43629 integral membrane protein from Beta vulgaris
and is a member of the sugar transporter family
PF|00083. ES [Oryza sativa]
Length = 501
Score = 212 bits (540), Expect = 4e-54
Identities = 98/152 (64%), Positives = 128/152 (83%)
Frame = -2
Query: 737 STWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMV 558
+TWL DK+GRRLLL++S++ MT++LVVVS++F+++ ++ SHLY ++ ++S+VGLV V
Sbjct: 350 TTWLTDKAGRRLLLIISTTGMTITLVVVSVSFFVKDNITNGSHLYSVMSMLSLVGLVAFV 409
Query: 557 IGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGGTFTM 378
I FSLGLG IPW+IMSEILPV+IK LAGS AT+ANWLT+W ITMTA+L+L+WS+GGTF +
Sbjct: 410 ISFSLGLGAIPWIIMSEILPVNIKSLAGSVATLANWLTAWLITMTASLMLSWSNGGTFAI 469
Query: 377 YTVVAAFTVVFTALWVPETKGRTLEEIQSSFR 282
Y V A T+VF LWVPETKGRTLEEI SFR
Sbjct: 470 YAAVCAGTLVFVCLWVPETKGRTLEEIAFSFR 501
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 656,880,937
Number of Sequences: 1393205
Number of extensions: 15047536
Number of successful extensions: 41141
Number of sequences better than 10.0: 715
Number of HSP's better than 10.0 without gapping: 38006
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40421
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 35188080875
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)