Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000165A_C01 KMC000165A_c01
(677 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
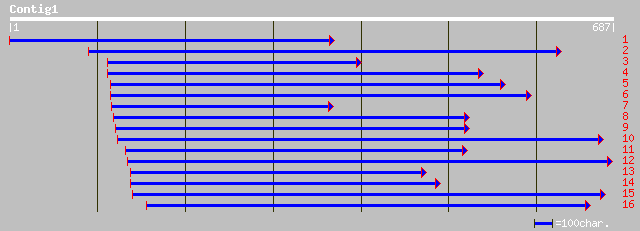
ref|NP_178216.1| transfactor-like protein; protein id: At2g01060... 86 4e-16
sp|P37800|RRPL_TOSV RNA-directed RNA polymerase (L protein) gi|3... 39 0.065
gb|AAB25907.1| large structural protein L=RNA-dependent RNA poly... 39 0.065
emb|CAB83215.1| XLalphas protein [Homo sapiens] gi|10086224|emb|... 34 2.1
ref|NP_536350.1| guanine nucleotide binding protein (G protein),... 34 2.1
>ref|NP_178216.1| transfactor-like protein; protein id: At2g01060.1, supported by
cDNA: gi_13899110 [Arabidopsis thaliana]
gi|25410874|pir||B84420 transfactor-like protein
[imported] - Arabidopsis thaliana
gi|6598621|gb|AAF18654.1| transfactor-like protein
[Arabidopsis thaliana]
gi|13899111|gb|AAK48977.1|AF370550_1 transfactor-like
protein [Arabidopsis thaliana]
gi|21536958|gb|AAM61299.1| transfactor-like protein
[Arabidopsis thaliana] gi|23197610|gb|AAN15332.1|
transfactor-like protein [Arabidopsis thaliana]
Length = 286
Score = 86.3 bits (212), Expect = 4e-16
Identities = 52/116 (44%), Positives = 69/116 (58%), Gaps = 2/116 (1%)
Frame = -1
Query: 674 TPDPEKAAKQHAPAKSLSI-ESFSSHHEPMTPDSACHVGSPADSPKEERSAKKQRVSMDG 498
+P +K+ K P KSLS+ ES SS+ EP+TPDS C++GSP +S EER +KK R+
Sbjct: 174 SPLQDKSGKDCGPDKSLSVDESLSSYREPLTPDSGCNIGSPDESTGEERLSKKPRLVRGA 233
Query: 497 AYSKPDIVLPHQILESSM-PSYQQPTTVFLTQEHFDSSLGISTRSEELEKIGGSNL 333
A PDIV+ H ILES + SY Q V + S LG E+L+K+ G NL
Sbjct: 234 AGYTPDIVVGHPILESGLNTSYHQSDHVLAFDQPSTSLLG---AEEQLDKVSGDNL 286
>sp|P37800|RRPL_TOSV RNA-directed RNA polymerase (L protein) gi|321509|pir||S29529
genome polyprotein - Toscana virus
gi|62200|emb|CAA48478.1| RNA-dependant RNA polymerase
[Toscana virus]
Length = 2095
Score = 38.9 bits (89), Expect = 0.065
Identities = 22/67 (32%), Positives = 33/67 (48%)
Frame = -1
Query: 539 EERSAKKQRVSMDGAYSKPDIVLPHQILESSMPSYQQPTTVFLTQEHFDSSLGISTRSEE 360
EE ++ V M+G S P+I PH++L + VFLT F + IS +
Sbjct: 666 EELQTMQRYVIMEGFVSLPEIPKPHKMLSKIPKVLRSELQVFLTHRLFSTMQRISATPFQ 725
Query: 359 LEKIGGS 339
L K+GG+
Sbjct: 726 LHKVGGN 732
>gb|AAB25907.1| large structural protein L=RNA-dependent RNA polymerase homolog
[Toscana virus TOS, Peptide, 2095 aa]
Length = 2095
Score = 38.9 bits (89), Expect = 0.065
Identities = 22/67 (32%), Positives = 33/67 (48%)
Frame = -1
Query: 539 EERSAKKQRVSMDGAYSKPDIVLPHQILESSMPSYQQPTTVFLTQEHFDSSLGISTRSEE 360
EE ++ V M+G S P+I PH++L + VFLT F + IS +
Sbjct: 666 EELQTMQRYVIMEGFVSLPEIPKPHKMLSKIPKVLRSELQVFLTHRLFSTMQRISATPFQ 725
Query: 359 LEKIGGS 339
L K+GG+
Sbjct: 726 LHKVGGN 732
>emb|CAB83215.1| XLalphas protein [Homo sapiens] gi|10086224|emb|CAC07997.1|
dJ806M20.3.1 (isoform 1 of guanine nucleotide binding
protein (G protein), alpha stimulating activity
polypeptide 1) [Homo sapiens]
Length = 388
Score = 33.9 bits (76), Expect = 2.1
Identities = 28/104 (26%), Positives = 48/104 (45%), Gaps = 5/104 (4%)
Frame = -1
Query: 677 ATPDPEKAAKQHAPAKSLSIESFSSHHEPMTPDSACHVGSP-ADSPKEERSAKKQRVS-- 507
A DP+ A APA + E+ ++H P PD+ G+P A + R+A+ +R +
Sbjct: 160 APADPDAGAAPEAPAAPAAAETRAAHVAPAAPDA----GAPTAPAASATRAAQVRRAASA 215
Query: 506 --MDGAYSKPDIVLPHQILESSMPSYQQPTTVFLTQEHFDSSLG 381
GA K + P ++++ P +PT + +SS G
Sbjct: 216 APASGARRKIHLRPPSPEIQAADPPTPRPTRASAWRGKSESSRG 259
>ref|NP_536350.1| guanine nucleotide binding protein (G protein), alpha stimulating
activity polypeptide 1, isoform XL-alpha-s; GNAS complex
locus; neuroendocrine secretory protein 55; guanine
nucleotide regulatory protein; G-s-alpha; paternally
expressed G protein XL alpha S [Homo sapiens]
Length = 909
Score = 33.9 bits (76), Expect = 2.1
Identities = 28/104 (26%), Positives = 48/104 (45%), Gaps = 5/104 (4%)
Frame = -1
Query: 677 ATPDPEKAAKQHAPAKSLSIESFSSHHEPMTPDSACHVGSP-ADSPKEERSAKKQRVS-- 507
A DP+ A APA + E+ ++H P PD+ G+P A + R+A+ +R +
Sbjct: 333 APADPDAGAAPEAPAAPAAAETRAAHVAPAAPDA----GAPTAPAASATRAAQVRRAASA 388
Query: 506 --MDGAYSKPDIVLPHQILESSMPSYQQPTTVFLTQEHFDSSLG 381
GA K + P ++++ P +PT + +SS G
Sbjct: 389 APASGARRKIHLRPPSPEIQAADPPTPRPTRASAWRGKSESSRG 432
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 571,103,437
Number of Sequences: 1393205
Number of extensions: 12000660
Number of successful extensions: 32936
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 31105
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32783
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29987172312
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)