Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000158A_C01 KMC000158A_c01
(557 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
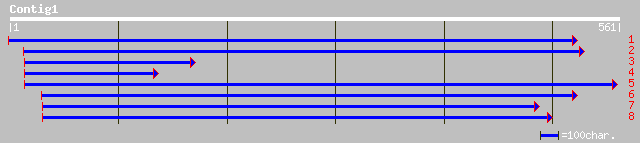
ref|NP_190904.1| putative protein; protein id: At3g53360.1 [Arab... 69 1e-11
pir||F71401 hypothetical protein - Arabidopsis thaliana 43 0.002
ref|NP_193141.2| hypothetical protein; protein id: At4g14050.1 [... 43 0.002
pir||B85153 hypothetical protein AT4g14050 [imported] - Arabidop... 43 0.002
ref|NP_193809.1| putative protein; protein id: At4g20770.1 [Arab... 40 0.019
>ref|NP_190904.1| putative protein; protein id: At3g53360.1 [Arabidopsis thaliana]
gi|11281375|pir||T45876 hypothetical protein F4P12.60 -
Arabidopsis thaliana gi|6729487|emb|CAB67643.1| putative
protein [Arabidopsis thaliana]
Length = 768
Score = 69.3 bits (168), Expect(2) = 1e-11
Identities = 46/121 (38%), Positives = 64/121 (52%), Gaps = 9/121 (7%)
Frame = -1
Query: 428 SMYTNFGPIAHALYMFTMICINDIIFWGSMITGFSQLYSEI--------CQGTGVCQPNE 273
+MY F ++ A +F I + D+I W S+I GFSQL E GV PNE
Sbjct: 211 AMYVRFNQMSDASRVFYGIPMKDLISWSSIIAGFSQLGFEFEALSHLKEMLSFGVFHPNE 270
Query: 272 FIFGSVFSRLSACCSLLEPEY-EGIYMEYVLNFI*GGTLSPNTLCYMYMKFGFLPSAKMT 96
+IFG S L AC SLL P+Y I+ + + + G ++ +LC MY + GFL SA+
Sbjct: 271 YIFG---SSLKACSSLLRPDYGSQIHGLCIKSELAGNAIAGCSLCDMYARCGFLNSARRV 327
Query: 95 F 93
F
Sbjct: 328 F 328
Score = 35.0 bits (79), Expect = 0.62
Identities = 31/122 (25%), Positives = 57/122 (46%), Gaps = 10/122 (8%)
Frame = -1
Query: 428 SMYTNFGPIAHALYMFTMICINDIIFWGSMITGFSQ---------LYSEICQGTGVCQPN 276
SMY G + A +F + +++ + S+ITG+SQ LY ++ Q V P+
Sbjct: 110 SMYGKCGSLRDAREVFDFMPERNLVSYTSVITGYSQNGQGAEAIRLYLKMLQEDLV--PD 167
Query: 275 EFIFGSVFSRLSACCSLLEPEY-EGIYMEYVLNFI*GGTLSPNTLCYMYMKFGFLPSAKM 99
+F FGS+ + AC S + + ++ + + ++ N L MY++F + A
Sbjct: 168 QFAFGSI---IKACASSSDVGLGKQLHAQVIKLESSSHLIAQNALIAMYVRFNQMSDASR 224
Query: 98 TF 93
F
Sbjct: 225 VF 226
Score = 21.2 bits (43), Expect(2) = 1e-11
Identities = 9/24 (37%), Positives = 15/24 (62%)
Frame = -2
Query: 502 IDRKLHVHVHKSGFNCHLIAHSAI 431
+ ++LH V K + HLIA +A+
Sbjct: 186 LGKQLHAQVIKLESSSHLIAQNAL 209
>pir||F71401 hypothetical protein - Arabidopsis thaliana
Length = 258
Score = 43.1 bits (100), Expect = 0.002
Identities = 30/122 (24%), Positives = 62/122 (50%), Gaps = 10/122 (8%)
Frame = -1
Query: 428 SMYTNFGPIAHALYMFTMICINDIIFWGSMITGFSQ---------LYSEICQGTGVCQPN 276
++Y G +HAL +F + D I W S++T +Q ++S + +G+ +P+
Sbjct: 114 NVYGKCGAASHALQVFDEMPHRDHIAWASVLTALNQANLSGKTLSVFSSVGSSSGL-RPD 172
Query: 275 EFIFGSVFSRLSACCSLLEPEY-EGIYMEYVLNFI*GGTLSPNTLCYMYMKFGFLPSAKM 99
+F+F ++ + AC +L ++ ++ ++++ + ++L MY K G L SAK
Sbjct: 173 DFVFSAL---VKACANLGSIDHGRQVHCHFIVSEYANDEVVKSSLVDMYAKCGLLNSAKA 229
Query: 98 TF 93
F
Sbjct: 230 VF 231
>ref|NP_193141.2| hypothetical protein; protein id: At4g14050.1 [Arabidopsis
thaliana]
Length = 612
Score = 43.1 bits (100), Expect = 0.002
Identities = 30/122 (24%), Positives = 62/122 (50%), Gaps = 10/122 (8%)
Frame = -1
Query: 428 SMYTNFGPIAHALYMFTMICINDIIFWGSMITGFSQ---------LYSEICQGTGVCQPN 276
++Y G +HAL +F + D I W S++T +Q ++S + +G+ +P+
Sbjct: 46 NVYGKCGAASHALQVFDEMPHRDHIAWASVLTALNQANLSGKTLSVFSSVGSSSGL-RPD 104
Query: 275 EFIFGSVFSRLSACCSLLEPEY-EGIYMEYVLNFI*GGTLSPNTLCYMYMKFGFLPSAKM 99
+F+F ++ + AC +L ++ ++ ++++ + ++L MY K G L SAK
Sbjct: 105 DFVFSAL---VKACANLGSIDHGRQVHCHFIVSEYANDEVVKSSLVDMYAKCGLLNSAKA 161
Query: 98 TF 93
F
Sbjct: 162 VF 163
>pir||B85153 hypothetical protein AT4g14050 [imported] - Arabidopsis thaliana
gi|5280987|emb|CAB46001.1| hypothetical protein
[Arabidopsis thaliana] gi|7268109|emb|CAB78447.1|
hypothetical protein [Arabidopsis thaliana]
Length = 686
Score = 43.1 bits (100), Expect = 0.002
Identities = 30/122 (24%), Positives = 62/122 (50%), Gaps = 10/122 (8%)
Frame = -1
Query: 428 SMYTNFGPIAHALYMFTMICINDIIFWGSMITGFSQ---------LYSEICQGTGVCQPN 276
++Y G +HAL +F + D I W S++T +Q ++S + +G+ +P+
Sbjct: 114 NVYGKCGAASHALQVFDEMPHRDHIAWASVLTALNQANLSGKTLSVFSSVGSSSGL-RPD 172
Query: 275 EFIFGSVFSRLSACCSLLEPEY-EGIYMEYVLNFI*GGTLSPNTLCYMYMKFGFLPSAKM 99
+F+F ++ + AC +L ++ ++ ++++ + ++L MY K G L SAK
Sbjct: 173 DFVFSAL---VKACANLGSIDHGRQVHCHFIVSEYANDEVVKSSLVDMYAKCGLLNSAKA 229
Query: 98 TF 93
F
Sbjct: 230 VF 231
>ref|NP_193809.1| putative protein; protein id: At4g20770.1 [Arabidopsis thaliana]
gi|7485927|pir||T10619 hypothetical protein F21C20.120 -
Arabidopsis thaliana gi|5262217|emb|CAB45843.1| putative
protein [Arabidopsis thaliana]
gi|7268873|emb|CAB79077.1| putative protein [Arabidopsis
thaliana]
Length = 740
Score = 40.0 bits (92), Expect = 0.019
Identities = 36/107 (33%), Positives = 49/107 (45%), Gaps = 14/107 (13%)
Frame = -1
Query: 371 CIN--DIIFWGSMITGFSQ---------LYSEICQGTGVCQPNEFIFGSVFSRLSACCSL 225
CIN DI W SMI+GF L+ + Q +C PNE F +V S S CSL
Sbjct: 443 CINELDIACWNSMISGFRHNMLDTKALILFRRMHQTAVLC-PNETSFATVLSSCSRLCSL 501
Query: 224 LE-PEYEGIYME--YVLNFI*GGTLSPNTLCYMYMKFGFLPSAKMTF 93
L ++ G+ ++ YV + L MY K G + SA+ F
Sbjct: 502 LHGRQFHGLVVKSGYV-----SDSFVETALTDMYCKCGEIDSARQFF 543
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 471,938,128
Number of Sequences: 1393205
Number of extensions: 10306155
Number of successful extensions: 23506
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 22217
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23478
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)