Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000151A_C01 KMC000151A_c01
(677 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
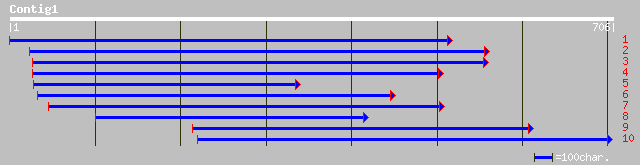
ref|NP_187193.1| unknown protein; protein id: At3g05420.1 [Arabi... 167 2e-40
ref|NP_198115.2| putative protein; protein id: At5g27630.1, supp... 152 3e-36
ref|NP_196062.1| putative protein; protein id: At5g04420.1, supp... 90 3e-17
gb|AAL06496.1|AF412043_1 AT5g04420/T32M21_20 [Arabidopsis thaliana] 90 3e-17
gb|EAA37369.1| GLP_24_16856_21838 [Giardia lamblia ATCC 50803] 45 0.001
>ref|NP_187193.1| unknown protein; protein id: At3g05420.1 [Arabidopsis thaliana]
gi|7596769|gb|AAF64540.1| unknown protein [Arabidopsis
thaliana]
Length = 668
Score = 167 bits (422), Expect = 2e-40
Identities = 86/110 (78%), Positives = 100/110 (90%)
Frame = -1
Query: 677 ESSLSKEKLHTLQLKQTLGEAEGRNSDLCKELQSVRGQLAAEQSRCFKLEVEVAELGQKL 498
E+SL+KE++ TLQL+Q LGEAE RN+DL KELQSVRGQLAAEQSRCFKLEV+VAEL QKL
Sbjct: 558 EASLNKERMQTLQLRQELGEAELRNTDLYKELQSVRGQLAAEQSRCFKLEVDVAELRQKL 617
Query: 497 QTIGTLQKELELLQRQKAASEQAALSAKPRQGSGGVWGWLAGTPPNQQEE 348
QT+ TLQKELELLQRQKAASEQAA++AK RQGSGGVWGWLAG+P + ++
Sbjct: 618 QTLETLQKELELLQRQKAASEQAAMNAK-RQGSGGVWGWLAGSPQEKDDD 666
>ref|NP_198115.2| putative protein; protein id: At5g27630.1, supported by cDNA:
gi_20260513 [Arabidopsis thaliana]
gi|20260514|gb|AAM13155.1| unknown protein [Arabidopsis
thaliana]
Length = 648
Score = 152 bits (385), Expect = 3e-36
Identities = 81/106 (76%), Positives = 90/106 (84%), Gaps = 1/106 (0%)
Frame = -1
Query: 677 ESSLSKEKLHTLQLKQTLGEAEGRNSDLCKELQSVRGQLAAEQSRCFKLEVEVAELGQKL 498
E+SL+KEK+ TLQLK+ L E + RN++L KELQSVR QLAAEQSRCFKLEVEVAEL QKL
Sbjct: 540 EASLNKEKIQTLQLKEELAEIDTRNTELYKELQSVRNQLAAEQSRCFKLEVEVAELRQKL 599
Query: 497 QTIGTLQKELELLQRQKA-ASEQAALSAKPRQGSGGVWGWLAGTPP 363
QT+ TLQKELELLQRQ+A ASEQAA RQ SGGVWGWLAGTPP
Sbjct: 600 QTMETLQKELELLQRQRAVASEQAATMNAKRQSSGGVWGWLAGTPP 645
>ref|NP_196062.1| putative protein; protein id: At5g04420.1, supported by cDNA:
gi_15724205 [Arabidopsis thaliana]
gi|11358331|pir||T48438 hypothetical protein T32M21.20 -
Arabidopsis thaliana gi|7406446|emb|CAB85548.1| putative
protein [Arabidopsis thaliana]
Length = 514
Score = 89.7 bits (221), Expect = 3e-17
Identities = 41/98 (41%), Positives = 71/98 (71%)
Frame = -1
Query: 677 ESSLSKEKLHTLQLKQTLGEAEGRNSDLCKELQSVRGQLAAEQSRCFKLEVEVAELGQKL 498
ESS+++ ++ +L++ + E +++L +ELQSV GQL +E+SRCFKLE ++AEL + L
Sbjct: 413 ESSIAETQVENAKLREKIDEVNSSHTELSQELQSVEGQLISERSRCFKLEAQIAELQKAL 472
Query: 497 QTIGTLQKELELLQRQKAASEQAALSAKPRQGSGGVWG 384
++ +++ E+E+L+RQ++AS++ RQGS GVWG
Sbjct: 473 ESGQSIEAEVEMLRRQRSASDEEEDGTVQRQGSAGVWG 510
>gb|AAL06496.1|AF412043_1 AT5g04420/T32M21_20 [Arabidopsis thaliana]
Length = 514
Score = 89.7 bits (221), Expect = 3e-17
Identities = 41/98 (41%), Positives = 71/98 (71%)
Frame = -1
Query: 677 ESSLSKEKLHTLQLKQTLGEAEGRNSDLCKELQSVRGQLAAEQSRCFKLEVEVAELGQKL 498
ESS+++ ++ +L++ + E +++L +ELQSV GQL +E+SRCFKLE ++AEL + L
Sbjct: 413 ESSIAETQVENAKLREKIDEVNSSHTELSQELQSVEGQLISERSRCFKLEAQIAELQKAL 472
Query: 497 QTIGTLQKELELLQRQKAASEQAALSAKPRQGSGGVWG 384
++ +++ E+E+L+RQ++AS++ RQGS GVWG
Sbjct: 473 ESGQSIEAEVEMLRRQRSASDEEEDGTVQRQGSAGVWG 510
>gb|EAA37369.1| GLP_24_16856_21838 [Giardia lamblia ATCC 50803]
Length = 1660
Score = 44.7 bits (104), Expect = 0.001
Identities = 30/82 (36%), Positives = 43/82 (51%)
Frame = -1
Query: 662 KEKLHTLQLKQTLGEAEGRNSDLCKELQSVRGQLAAEQSRCFKLEVEVAELGQKLQTIGT 483
K L LKQ E +G+N ++ EL+S + QL E SR +LE E++ + +LQ
Sbjct: 1340 KRNAELLDLKQQKDELKGKNYEISIELESAKRQLELETSRGLQLERELSNITSELQVARR 1399
Query: 482 LQKELELLQRQKAASEQAALSA 417
Q EL Q A+E AL+A
Sbjct: 1400 EQLELRTSASQ-LANENLALTA 1420
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 558,928,549
Number of Sequences: 1393205
Number of extensions: 11998514
Number of successful extensions: 53905
Number of sequences better than 10.0: 109
Number of HSP's better than 10.0 without gapping: 51694
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 53785
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29987172312
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)