Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000140A_C02 KMC000140A_c02
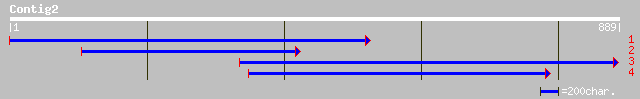
(889 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_610521.1| CG1663-PA [Drosophila melanogaster] gi|7303849|... 37 0.52
gb|EAA15183.1| hypothetical protein [Plasmodium yoelii yoelii] 35 1.5
ref|NP_116279.2| dispatched A [Homo sapiens] gi|27477708|ref|XP_... 35 2.0
pir||T42693 hypothetical protein DKFZp434I0428.1 - human (fragme... 35 2.0
dbj|BAB15365.1| unnamed protein product [Homo sapiens] 35 2.0
>ref|NP_610521.1| CG1663-PA [Drosophila melanogaster] gi|7303849|gb|AAF58895.1|
CG1663-PA [Drosophila melanogaster]
Length = 387
Score = 36.6 bits (83), Expect = 0.52
Identities = 22/61 (36%), Positives = 31/61 (50%), Gaps = 1/61 (1%)
Frame = -1
Query: 568 FHLYLLHPVVRVTIQFLRSNWIEWSNLFPYSHCFFSCLFPDY-HWLCNILSSNHVSVSIG 392
FH L VV + +FL+ W E + S F C +P Y H L + +NH+SV+I
Sbjct: 143 FHFQLDPKVVNASARFLQV-WFERQYVMQLSSSDFRCRYPKYYHSLLKFMPTNHISVTIC 201
Query: 391 E 389
E
Sbjct: 202 E 202
>gb|EAA15183.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 1860
Score = 35.0 bits (79), Expect = 1.5
Identities = 21/67 (31%), Positives = 35/67 (51%), Gaps = 4/67 (5%)
Frame = +1
Query: 406 LHGYSTEYCITNDNQEINKKRNNENMERD---CSIQSNCFSKIELSLSLLDAKGKDENC- 573
+H YS C NDN N R NE + CS +N + IE + + +++G + NC
Sbjct: 370 MHNYSYTDCYKNDNIIENSHRKNETNGTNINTCSTGNNIYKNIEKNNNNNNSRGFNHNCL 429
Query: 574 NASNSSL 594
N+S+ ++
Sbjct: 430 NSSDDNI 436
>ref|NP_116279.2| dispatched A [Homo sapiens] gi|27477708|ref|XP_209236.1| similar to
dispatched homolog 1; dispatched A; twelve-pass
transmembrane protein [Mus musculus] [Homo sapiens]
Length = 1524
Score = 34.7 bits (78), Expect = 2.0
Identities = 18/57 (31%), Positives = 30/57 (52%)
Frame = +1
Query: 439 NDNQEINKKRNNENMERDCSIQSNCFSKIELSLSLLDAKGKDENCNASNSSLLYFNL 609
N +E+ K R+ N+E ++ K+ELSLS DA E+ N + +L+ +L
Sbjct: 1411 NKQRELCKNRDVSNLESSGGTENKAGGKVELSLSQTDASVNSEHFNQNEPKVLFNHL 1467
>pir||T42693 hypothetical protein DKFZp434I0428.1 - human (fragment)
gi|6453545|emb|CAB61406.1| hypothetical protein [Homo
sapiens]
Length = 757
Score = 34.7 bits (78), Expect = 2.0
Identities = 18/57 (31%), Positives = 30/57 (52%)
Frame = +1
Query: 439 NDNQEINKKRNNENMERDCSIQSNCFSKIELSLSLLDAKGKDENCNASNSSLLYFNL 609
N +E+ K R+ N+E ++ K+ELSLS DA E+ N + +L+ +L
Sbjct: 644 NKQRELCKNRDVSNLESSGGTENKAGGKVELSLSQTDASVNSEHFNQNEPKVLFNHL 700
>dbj|BAB15365.1| unnamed protein product [Homo sapiens]
Length = 917
Score = 34.7 bits (78), Expect = 2.0
Identities = 18/57 (31%), Positives = 30/57 (52%)
Frame = +1
Query: 439 NDNQEINKKRNNENMERDCSIQSNCFSKIELSLSLLDAKGKDENCNASNSSLLYFNL 609
N +E+ K R+ N+E ++ K+ELSLS DA E+ N + +L+ +L
Sbjct: 804 NKQRELCKNRDVSNLESSGGTENKAGGKVELSLSQTDASVNSEHFNQNEPKVLFNHL 860
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 688,314,867
Number of Sequences: 1393205
Number of extensions: 14317462
Number of successful extensions: 40381
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 35803
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40054
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 48218255001
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)