Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000127A_C01 KMC000127A_c01
(997 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
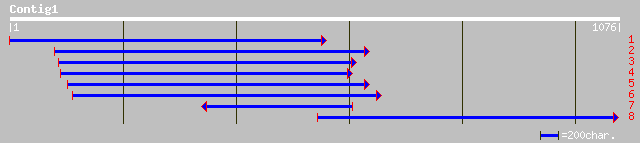
sp|P26969|GCSP_PEA Glycine dehydrogenase [decarboxylating], mito... 484 e-136
ref|NP_180178.1| putative glycine dehydrogenase; protein id: At2... 421 e-117
sp|O49954|GCSP_SOLTU Glycine dehydrogenase [decarboxylating], mi... 416 e-115
sp|P49361|GCSA_FLAPR Glycine dehydrogenase [decarboxylating] A, ... 412 e-114
gb|AAN17423.1| P-Protein - like protein [Arabidopsis thaliana] 410 e-113
>sp|P26969|GCSP_PEA Glycine dehydrogenase [decarboxylating], mitochondrial precursor
(Glycine decarboxylase) (Glycine cleavage system
P-protein) gi|282926|pir||A42109 glycine dehydrogenase
(decarboxylating) (EC 1.4.4.2) component P precursor -
garden pea gi|20741|emb|CAA42443.1| P protein; component
of aminomethyltransferase [Pisum sativum]
Length = 1057
Score = 484 bits (1247), Expect = e-136
Identities = 250/300 (83%), Positives = 268/300 (89%), Gaps = 7/300 (2%)
Frame = +2
Query: 119 MERARRLANRAILKRLVSEAKHNRKHEPVWNSAAASTTTVPF-YSASSSRYVSSVSHSVL 295
MERARRLANRA LKRL+SEAK NRK E +++ +TT +PF S SSSRYVSSVS+S+L
Sbjct: 1 MERARRLANRATLKRLLSEAKQNRKTE---STSTTTTTPLPFSLSGSSSRYVSSVSNSIL 57
Query: 296 RNRGSKSATNIPRAAAGL------SQTRSISVEALQPSDTFPRRHNSATPEEQAKMSLAC 457
R RGSK N+ R G SQ+RSISVEAL+PSDTFPRRHNSATP+EQ KM+ +
Sbjct: 58 RGRGSKPDNNVSRRVGGFLGVGYPSQSRSISVEALKPSDTFPRRHNSATPDEQTKMAESV 117
Query: 458 GFDNVDSLVDATVPKSIRLKEMKFNKFDGGLTEGQMIEHMKELASKNKVFKSFIGMGYYN 637
GFD +DSLVDATVPKSIRLKEMKFNKFDGGLTEGQMIEHMK+LASKNKVFKSFIGMGYYN
Sbjct: 118 GFDTLDSLVDATVPKSIRLKEMKFNKFDGGLTEGQMIEHMKDLASKNKVFKSFIGMGYYN 177
Query: 638 THVPPVILRNIMENPAWYTQYTPYQAEISQGRLESLLNYQTMITDLTGLPMSNASLLDEG 817
THVPPVILRNIMENPAWYTQYTPYQAEISQGRLESLLN+QTMITDLTGLPMSNASLLDEG
Sbjct: 178 THVPPVILRNIMENPAWYTQYTPYQAEISQGRLESLLNFQTMITDLTGLPMSNASLLDEG 237
Query: 818 TAAAEAMSMCNNIQKGKKKTFIIASNCHPQTIDICKTRADGFELKVVVADLKDIDYKSGD 997
TAAAEAMSMCNNIQKGKKKTFIIASNCHPQTIDIC+TRADGFELKVVV DLKDIDYKSGD
Sbjct: 238 TAAAEAMSMCNNIQKGKKKTFIIASNCHPQTIDICQTRADGFELKVVVKDLKDIDYKSGD 297
>ref|NP_180178.1| putative glycine dehydrogenase; protein id: At2g26080.1
[Arabidopsis thaliana] gi|12229797|sp|O80988|GCSP_ARATH
Glycine dehydrogenase [decarboxylating], mitochondrial
precursor (Glycine decarboxylase) (Glycine cleavage
system P-protein) gi|7488117|pir||T02615 probable
glycine dehydrogenase (decarboxylating) (EC 1.4.4.2)
T19L18.11 - Arabidopsis thaliana
gi|3413705|gb|AAC31228.1| putative glycine dehydrogenase
[Arabidopsis thaliana]
Length = 1044
Score = 421 bits (1082), Expect = e-117
Identities = 219/294 (74%), Positives = 245/294 (82%), Gaps = 1/294 (0%)
Frame = +2
Query: 119 MERARRLANRAILKRLVSEAKHNRKHEPVWNSAAASTTTVPFYSASSSRYVSSVSHSVLR 298
MERARRLA R I+KRLV+E K +R E S+ TTTV + SRYVSSVS S L
Sbjct: 1 MERARRLAYRGIVKRLVNETKRHRNGE----SSLLPTTTV-----TPSRYVSSVS-SFLH 50
Query: 299 NRGSKSATNIPRAAAGLSQTRSISVEALQPSDTFPRRHNSATPEEQAKMSLACGFDNVDS 478
R S + + QTRSISV+AL+PSDTFPRRHNSATP+EQA+M+ CGFDN+++
Sbjct: 51 RRRDVSGSAFTTSGRNQHQTRSISVDALKPSDTFPRRHNSATPDEQAQMANYCGFDNLNT 110
Query: 479 LVDATVPKSIRLKEMKFNK-FDGGLTEGQMIEHMKELASKNKVFKSFIGMGYYNTHVPPV 655
L+D+TVPKSIRL MKF+ FD GLTE QMIEHM +LASKNKVFKSFIGMGYYNTHVPPV
Sbjct: 111 LIDSTVPKSIRLDSMKFSGIFDEGLTESQMIEHMSDLASKNKVFKSFIGMGYYNTHVPPV 170
Query: 656 ILRNIMENPAWYTQYTPYQAEISQGRLESLLNYQTMITDLTGLPMSNASLLDEGTAAAEA 835
ILRNIMENPAWYTQYTPYQAEISQGRLESLLNYQT+ITDLTGLPMSNASLLDEGTAAAEA
Sbjct: 171 ILRNIMENPAWYTQYTPYQAEISQGRLESLLNYQTVITDLTGLPMSNASLLDEGTAAAEA 230
Query: 836 MSMCNNIQKGKKKTFIIASNCHPQTIDICKTRADGFELKVVVADLKDIDYKSGD 997
M+MCNNI KGKKKTF+IASNCHPQTID+CKTRADGF+LKVV D+KD+DY SGD
Sbjct: 231 MAMCNNILKGKKKTFVIASNCHPQTIDVCKTRADGFDLKVVTVDIKDVDYSSGD 284
>sp|O49954|GCSP_SOLTU Glycine dehydrogenase [decarboxylating], mitochondrial precursor
(Glycine decarboxylase) (Glycine cleavage system
P-protein) gi|7489225|pir||T07826 aminomethyltransferase
(EC 2.1.2.10) precursor - potato
gi|2894362|emb|CAB16918.1| P-Protein precursor [Solanum
tuberosum]
Length = 1035
Score = 416 bits (1068), Expect = e-115
Identities = 215/294 (73%), Positives = 241/294 (81%), Gaps = 1/294 (0%)
Frame = +2
Query: 119 MERARRLANRAILKRLVSEAKHNRKHEPVWNSAAASTTTVPFYSA-SSSRYVSSVSHSVL 295
MERAR+LANRAILKRLVS++K +R +E +P S SRYVSS+S
Sbjct: 1 MERARKLANRAILKRLVSQSKQSRSNE------------IPSSSLYRPSRYVSSLSPYTF 48
Query: 296 RNRGSKSATNIPRAAAGLSQTRSISVEALQPSDTFPRRHNSATPEEQAKMSLACGFDNVD 475
+ R + + N Q RSISVEAL+PSDTFPRRHNSATPEEQ KM+ CGF ++D
Sbjct: 49 QARNNAKSFNT-------QQARSISVEALKPSDTFPRRHNSATPEEQTKMAEFCGFQSLD 101
Query: 476 SLVDATVPKSIRLKEMKFNKFDGGLTEGQMIEHMKELASKNKVFKSFIGMGYYNTHVPPV 655
+L+DATVP+SIR + MK KFD GLTE QMIEHM+ LASKNKVFKS+IGMGYYNT+VPPV
Sbjct: 102 ALIDATVPQSIRSESMKLPKFDSGLTESQMIEHMQNLASKNKVFKSYIGMGYYNTYVPPV 161
Query: 656 ILRNIMENPAWYTQYTPYQAEISQGRLESLLNYQTMITDLTGLPMSNASLLDEGTAAAEA 835
ILRN++ENPAWYTQYTPYQAEISQGRLESLLNYQTMITDLTGLPMSNASLLDEGTAAAEA
Sbjct: 162 ILRNLLENPAWYTQYTPYQAEISQGRLESLLNYQTMITDLTGLPMSNASLLDEGTAAAEA 221
Query: 836 MSMCNNIQKGKKKTFIIASNCHPQTIDICKTRADGFELKVVVADLKDIDYKSGD 997
M+MCNNI KGKKKTF+IASNCHPQTIDICKTRADGF+LKVV DLKDIDYKSGD
Sbjct: 222 MAMCNNILKGKKKTFLIASNCHPQTIDICKTRADGFDLKVVTVDLKDIDYKSGD 275
>sp|P49361|GCSA_FLAPR Glycine dehydrogenase [decarboxylating] A, mitochondrial precursor
(Glycine decarboxylase A) (Glycine cleavage system
P-protein A) gi|2130011|pir||S63535
aminomethyltransferase (EC 2.1.2.10) gdcsPA precursor -
Flaveria pringlei gi|608712|emb|CAA85353.1| P-protein of
the glycine cleavage system [Flaveria pringlei]
Length = 1037
Score = 412 bits (1058), Expect = e-114
Identities = 216/293 (73%), Positives = 238/293 (80%)
Frame = +2
Query: 119 MERARRLANRAILKRLVSEAKHNRKHEPVWNSAAASTTTVPFYSASSSRYVSSVSHSVLR 298
MERARRLAN+AIL RLVS+ KHN P +S A S SRYVSS+S V
Sbjct: 1 MERARRLANKAILGRLVSQTKHN----PSISSPAL---------CSPSRYVSSLSPYVCS 47
Query: 299 NRGSKSATNIPRAAAGLSQTRSISVEALQPSDTFPRRHNSATPEEQAKMSLACGFDNVDS 478
+S N+ SQ R+ISVEAL+PSDTFPRRHNSATPEEQ KM+ GF N+DS
Sbjct: 48 GTNVRSDRNLNGFG---SQVRTISVEALKPSDTFPRRHNSATPEEQTKMAEFVGFPNLDS 104
Query: 479 LVDATVPKSIRLKEMKFNKFDGGLTEGQMIEHMKELASKNKVFKSFIGMGYYNTHVPPVI 658
L+DATVPKSIRL MK++KFD GLTE QMI HM++LASKNK+FKSFIGMGYYNT VP VI
Sbjct: 105 LIDATVPKSIRLDSMKYSKFDEGLTESQMIAHMQDLASKNKIFKSFIGMGYYNTSVPTVI 164
Query: 659 LRNIMENPAWYTQYTPYQAEISQGRLESLLNYQTMITDLTGLPMSNASLLDEGTAAAEAM 838
LRNIMENP WYTQYTPYQAEI+QGRLESLLN+QTM+TDLTGLPMSNASLLDEGTAAAEAM
Sbjct: 165 LRNIMENPGWYTQYTPYQAEIAQGRLESLLNFQTMVTDLTGLPMSNASLLDEGTAAAEAM 224
Query: 839 SMCNNIQKGKKKTFIIASNCHPQTIDICKTRADGFELKVVVADLKDIDYKSGD 997
+MCNNIQKGKKKTFIIASNCHPQTIDICKTRADGF+LKVV +DLKD DY SGD
Sbjct: 225 AMCNNIQKGKKKTFIIASNCHPQTIDICKTRADGFDLKVVTSDLKDFDYSSGD 277
>gb|AAN17423.1| P-Protein - like protein [Arabidopsis thaliana]
Length = 956
Score = 410 bits (1055), Expect = e-113
Identities = 211/293 (72%), Positives = 238/293 (81%)
Frame = +2
Query: 119 MERARRLANRAILKRLVSEAKHNRKHEPVWNSAAASTTTVPFYSASSSRYVSSVSHSVLR 298
MERARRLA R I+KRLV++ K +R E T + +RYVSS+S +
Sbjct: 1 MERARRLAYRGIVKRLVNDTKRHRNAE-----------TPHLVPHAPARYVSSLSPFIST 49
Query: 299 NRGSKSATNIPRAAAGLSQTRSISVEALQPSDTFPRRHNSATPEEQAKMSLACGFDNVDS 478
R + N A QTRSISV+A++PSDTFPRRHNSATP+EQ M+ CGFD++DS
Sbjct: 50 PR----SVNHTAAFGRHQQTRSISVDAVKPSDTFPRRHNSATPDEQTHMAKFCGFDHIDS 105
Query: 479 LVDATVPKSIRLKEMKFNKFDGGLTEGQMIEHMKELASKNKVFKSFIGMGYYNTHVPPVI 658
L+DATVPKSIRL MKF+KFD GLTE QMI+HM +LASKNKVFKSFIGMGYYNTHVP VI
Sbjct: 106 LIDATVPKSIRLDSMKFSKFDAGLTESQMIQHMVDLASKNKVFKSFIGMGYYNTHVPTVI 165
Query: 659 LRNIMENPAWYTQYTPYQAEISQGRLESLLNYQTMITDLTGLPMSNASLLDEGTAAAEAM 838
LRNIMENPAWYTQYTPYQAEISQGRLESLLN+QT+ITDLTGLPMSNASLLDEGTAAAEAM
Sbjct: 166 LRNIMENPAWYTQYTPYQAEISQGRLESLLNFQTVITDLTGLPMSNASLLDEGTAAAEAM 225
Query: 839 SMCNNIQKGKKKTFIIASNCHPQTIDICKTRADGFELKVVVADLKDIDYKSGD 997
+MCNNI KGKKKTF+IASNCHPQTID+CKTRADGF+LKVV +DLKDIDY SGD
Sbjct: 226 AMCNNILKGKKKTFVIASNCHPQTIDVCKTRADGFDLKVVTSDLKDIDYSSGD 278
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 945,003,599
Number of Sequences: 1393205
Number of extensions: 24119987
Number of successful extensions: 206949
Number of sequences better than 10.0: 1311
Number of HSP's better than 10.0 without gapping: 99096
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 150312
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 57117888189
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)