Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000125A_C01 KMC000125A_c01
(596 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
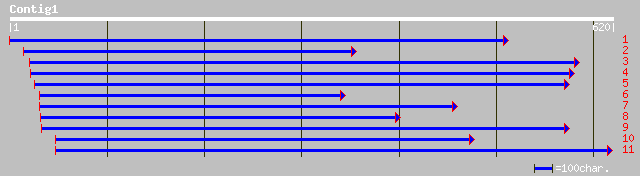
dbj|BAB70751.1| respiratory burst oxidase homolog [Solanum tuber... 155 5e-37
pir||B86223 hypothetical protein [imported] - Arabidopsis thalia... 152 4e-36
ref|NP_172383.1| putative respiratory burst oxidase protein B; p... 152 4e-36
gb|AAC39476.1| respiratory burst oxidase protein B [Arabidopsis ... 152 4e-36
gb|AAC39475.1| respiratory burst oxidase protein A [Arabidopsis ... 146 2e-34
>dbj|BAB70751.1| respiratory burst oxidase homolog [Solanum tuberosum]
Length = 867
Score = 155 bits (391), Expect = 5e-37
Identities = 71/107 (66%), Positives = 86/107 (80%)
Frame = -3
Query: 594 KEGVIELHDYCTGVYKEGDDGSALITMLQSLYRSQYGVDKVSETRVKTHFGRPNWRNVFE 415
+EG+IELH+YCT VY+EGD SALITMLQS+ +++ GVD VS TRVKTHF RPNWR VF+
Sbjct: 761 QEGLIELHNYCTSVYEEGDARSALITMLQSIQQAKSGVDIVSGTRVKTHFARPNWRQVFK 820
Query: 414 HVSLKHPDKRVGVFYCGAHGLIGELKRLSLDFSRKTSTKFEFSPREF 274
V++ HPD+R+GVFYCG GL+GEL+ LS DFS KT TKFEF F
Sbjct: 821 RVTINHPDQRIGVFYCGPQGLVGELRHLSQDFSHKTGTKFEFHKENF 867
>pir||B86223 hypothetical protein [imported] - Arabidopsis thaliana
gi|2342676|gb|AAB70398.1| Strong similarity to Oryza
NADPH oxidase (gb|X93301). [Arabidopsis thaliana]
Length = 578
Score = 152 bits (383), Expect = 4e-36
Identities = 71/106 (66%), Positives = 88/106 (82%)
Frame = -3
Query: 591 EGVIELHDYCTGVYKEGDDGSALITMLQSLYRSQYGVDKVSETRVKTHFGRPNWRNVFEH 412
EG+IELH+YCT VY+EGD SALITMLQSL+ ++ G+D VS TRV+THF RPNWR+VF+H
Sbjct: 473 EGMIELHNYCTSVYEEGDARSALITMLQSLHHAKSGIDIVSGTRVRTHFARPNWRSVFKH 532
Query: 411 VSLKHPDKRVGVFYCGAHGLIGELKRLSLDFSRKTSTKFEFSPREF 274
V++ H ++RVGVFYCG +IGELKRL+ DFSRKT+TKFEF F
Sbjct: 533 VAVNHVNQRVGVFYCGNTCIIGELKRLAQDFSRKTTTKFEFHKENF 578
>ref|NP_172383.1| putative respiratory burst oxidase protein B; protein id: At1g09090.1
[Arabidopsis thaliana]
Length = 838
Score = 152 bits (383), Expect = 4e-36
Identities = 71/106 (66%), Positives = 88/106 (82%)
Frame = -3
Query: 591 EGVIELHDYCTGVYKEGDDGSALITMLQSLYRSQYGVDKVSETRVKTHFGRPNWRNVFEH 412
EG+IELH+YCT VY+EGD SALITMLQSL+ ++ G+D VS TRV+THF RPNWR+VF+H
Sbjct: 733 EGMIELHNYCTSVYEEGDARSALITMLQSLHHAKSGIDIVSGTRVRTHFARPNWRSVFKH 792
Query: 411 VSLKHPDKRVGVFYCGAHGLIGELKRLSLDFSRKTSTKFEFSPREF 274
V++ H ++RVGVFYCG +IGELKRL+ DFSRKT+TKFEF F
Sbjct: 793 VAVNHVNQRVGVFYCGNTCIIGELKRLAQDFSRKTTTKFEFHKENF 838
>gb|AAC39476.1| respiratory burst oxidase protein B [Arabidopsis thaliana]
Length = 843
Score = 152 bits (383), Expect = 4e-36
Identities = 71/106 (66%), Positives = 88/106 (82%)
Frame = -3
Query: 591 EGVIELHDYCTGVYKEGDDGSALITMLQSLYRSQYGVDKVSETRVKTHFGRPNWRNVFEH 412
EG+IELH+YCT VY+EGD SALITMLQSL+ ++ G+D VS TRV+THF RPNWR+VF+H
Sbjct: 738 EGMIELHNYCTSVYEEGDARSALITMLQSLHHAKSGIDIVSGTRVRTHFARPNWRSVFKH 797
Query: 411 VSLKHPDKRVGVFYCGAHGLIGELKRLSLDFSRKTSTKFEFSPREF 274
V++ H ++RVGVFYCG +IGELKRL+ DFSRKT+TKFEF F
Sbjct: 798 VAVNHVNQRVGVFYCGNTCIIGELKRLAQDFSRKTTTKFEFHKENF 843
>gb|AAC39475.1| respiratory burst oxidase protein A [Arabidopsis thaliana]
Length = 902
Score = 146 bits (368), Expect = 2e-34
Identities = 69/107 (64%), Positives = 84/107 (78%)
Frame = -3
Query: 594 KEGVIELHDYCTGVYKEGDDGSALITMLQSLYRSQYGVDKVSETRVKTHFGRPNWRNVFE 415
++ VIELH+YCT VY+EGD SALITMLQSL +++GVD VS TRV +HF RPNWR+VF+
Sbjct: 796 RKNVIELHNYCTSVYEEGDARSALITMLQSLNHAKHGVDVVSGTRVMSHFARPNWRSVFK 855
Query: 414 HVSLKHPDKRVGVFYCGAHGLIGELKRLSLDFSRKTSTKFEFSPREF 274
+++ HP RVGVFYCGA GL+ EL+ LSLDFS KTSTKF F F
Sbjct: 856 RIAVNHPKTRVGVFYCGAAGLVKELRHLSLDFSHKTSTKFIFHKENF 902
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 484,661,028
Number of Sequences: 1393205
Number of extensions: 9950547
Number of successful extensions: 22629
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 22098
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22598
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23140425222
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)