Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000124A_C01 KMC000124A_c01
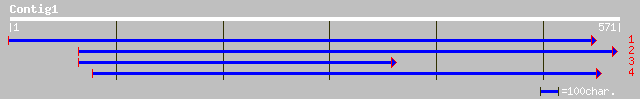
(571 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|EAA13496.1| agCP5216 [Anopheles gambiae str. PEST] 38 0.078
gb|AAL38986.1| cytochrome P450-3 [Musa acuminata] 35 0.66
ref|NP_650505.2| CG31183-PA [Drosophila melanogaster] gi|2317140... 32 7.3
gb|AAK92838.1| GH09326p [Drosophila melanogaster] 32 7.3
dbj|BAA88196.1| unnamed protein product [Oryza sativa (japonica ... 31 9.5
>gb|EAA13496.1| agCP5216 [Anopheles gambiae str. PEST]
Length = 456
Score = 38.1 bits (87), Expect = 0.078
Identities = 46/153 (30%), Positives = 64/153 (41%), Gaps = 17/153 (11%)
Frame = +2
Query: 128 NLLFGQ--AISVATCQRHTYICITTQLHYTILIYFEKNCSSRTSIMMKGTYLHPETDMRY 301
NLL Q A ++ T H IC LH + F K C+ R ++ T HP+ M +
Sbjct: 20 NLLASQVCAATMGTELAHCQICANCLLHVLRTLLFLKTCTPRLFMLHLPT--HPQ--MHF 75
Query: 302 VEEFQE*PTYKQIAQCSVPFPSTGPETLQHEYIL------------ISKLRLYLHSRSLS 445
V F+ KQ+ F S + + H +L +S LR +L RS S
Sbjct: 76 VPCFRSLIPLKQVDTIYTDFKSAF-DRIPHSLLLNKISQLDVNNLFVSWLRSFLCDRSYS 134
Query: 446 VNTN---KTPTSLSRFPPSRSQLRIPML*TVFL 535
V N TP S S P S L P+L +F+
Sbjct: 135 VKFNDMFSTPFSCSAGVPQGSVLS-PLLFIIFI 166
>gb|AAL38986.1| cytochrome P450-3 [Musa acuminata]
Length = 491
Score = 35.0 bits (79), Expect = 0.66
Identities = 26/94 (27%), Positives = 46/94 (48%), Gaps = 1/94 (1%)
Frame = +2
Query: 26 RERERERERESLHQGGNTILFSFFIQKDKKVSKL-NLLFGQAISVATCQRHTYICITTQL 202
RERERERERE +H G++ L + KK L +L FG+ V + + ++
Sbjct: 26 RERERERERERVHLLGSSSLHRSLWELSKKHGPLMHLKFGRVPVV--------VVSSPEM 77
Query: 203 HYTILIYFEKNCSSRTSIMMKGTYLHPETDMRYV 304
+L + C SR S++ + + +D+ ++
Sbjct: 78 AKEVLKTHDLECCSRPSLLSFSKFSYGLSDVAFI 111
>ref|NP_650505.2| CG31183-PA [Drosophila melanogaster] gi|23171402|gb|AAF55244.2|
CG31183-PA [Drosophila melanogaster]
Length = 1417
Score = 31.6 bits (70), Expect = 7.3
Identities = 17/39 (43%), Positives = 24/39 (60%), Gaps = 2/39 (5%)
Frame = +3
Query: 120 LSLIYFLVRQFQ*LHARDI--HIYVSQLNCIIQSLFTLK 230
LSL++ +VR Q LH+ DI H + NC++ S F LK
Sbjct: 697 LSLMHDIVRGMQFLHSSDIRSHGNLKSSNCVVDSRFVLK 735
>gb|AAK92838.1| GH09326p [Drosophila melanogaster]
Length = 821
Score = 31.6 bits (70), Expect = 7.3
Identities = 17/39 (43%), Positives = 24/39 (60%), Gaps = 2/39 (5%)
Frame = +3
Query: 120 LSLIYFLVRQFQ*LHARDI--HIYVSQLNCIIQSLFTLK 230
LSL++ +VR Q LH+ DI H + NC++ S F LK
Sbjct: 101 LSLMHDIVRGMQFLHSSDIRSHGNLKSSNCVVDSRFVLK 139
>dbj|BAA88196.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 965
Score = 31.2 bits (69), Expect = 9.5
Identities = 17/31 (54%), Positives = 19/31 (60%)
Frame = +3
Query: 27 ERERERERENLFIRGVILSYFLSLSKRTRKF 119
EREREREREN F RG + +S S R F
Sbjct: 9 ERERERERENAFRRGEQRAATISSSSREGHF 39
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 460,999,975
Number of Sequences: 1393205
Number of extensions: 9627432
Number of successful extensions: 27074
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 20182
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23942
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20956655091
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)