Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000120A_C01 KMC000120A_c01
(803 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
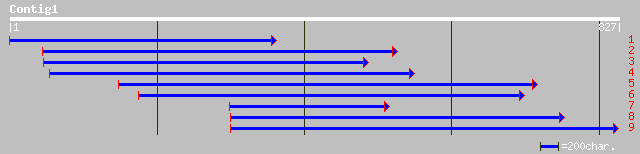
gb|AAL09731.1| At2g32240/F22D22.1 [Arabidopsis thaliana] gi|2026... 102 9e-21
ref|NP_565741.1| putative myosin heavy chain; protein id: At2g32... 102 9e-21
ref|NP_172024.1| unknown protein; protein id: At1g05320.1 [Arabi... 76 7e-13
pir||F84730 probable myosin heavy chain [imported] - Arabidopsis... 74 3e-12
ref|NP_341918.1| Hypothetical protein [Sulfolobus solfataricus] ... 41 0.018
>gb|AAL09731.1| At2g32240/F22D22.1 [Arabidopsis thaliana]
gi|20260364|gb|AAM13080.1| putative myosin heavy chain
[Arabidopsis thaliana] gi|22136178|gb|AAM91167.1|
putative myosin heavy chain [Arabidopsis thaliana]
Length = 568
Score = 102 bits (253), Expect = 9e-21
Identities = 60/127 (47%), Positives = 80/127 (62%), Gaps = 1/127 (0%)
Frame = -3
Query: 795 ELEASLKNSLEELEIKKNEITLLQKQVTDLEQKLQLTSDKLSVKGEVGVDQKDGLEVKSR 616
ELE++LK S EE+E KK +T + V DLEQK+QL K + V VKSR
Sbjct: 448 ELESALKKSQEEIEAKKKAVTEFESMVKDLEQKVQLADAKTKETEAMDVG------VKSR 501
Query: 615 DIGSSISSPSKRKSKKKSEATTAPTPSSSS-ETHVQTSQASPVINFKFILGVALVSIIFG 439
DI S SSP+KRKSKKK EA+ + + SS + T QT+ S ++ K + GVAL+S+I G
Sbjct: 502 DIDLSFSSPTKRKSKKKPEASLSSSSSSGNVTTPTQTASTSHLMTVKIVTGVALISVIIG 561
Query: 438 IILGKRY 418
IILG++Y
Sbjct: 562 IILGRKY 568
>ref|NP_565741.1| putative myosin heavy chain; protein id: At2g32240.1, supported by
cDNA: gi_15982766 [Arabidopsis thaliana]
gi|20197623|gb|AAM15156.1| putative myosin heavy chain
[Arabidopsis thaliana]
Length = 775
Score = 102 bits (253), Expect = 9e-21
Identities = 60/127 (47%), Positives = 80/127 (62%), Gaps = 1/127 (0%)
Frame = -3
Query: 795 ELEASLKNSLEELEIKKNEITLLQKQVTDLEQKLQLTSDKLSVKGEVGVDQKDGLEVKSR 616
ELE++LK S EE+E KK +T + V DLEQK+QL K + V VKSR
Sbjct: 655 ELESALKKSQEEIEAKKKAVTEFESMVKDLEQKVQLADAKTKETEAMDVG------VKSR 708
Query: 615 DIGSSISSPSKRKSKKKSEATTAPTPSSSS-ETHVQTSQASPVINFKFILGVALVSIIFG 439
DI S SSP+KRKSKKK EA+ + + SS + T QT+ S ++ K + GVAL+S+I G
Sbjct: 709 DIDLSFSSPTKRKSKKKPEASLSSSSSSGNVTTPTQTASTSHLMTVKIVTGVALISVIIG 768
Query: 438 IILGKRY 418
IILG++Y
Sbjct: 769 IILGRKY 775
>ref|NP_172024.1| unknown protein; protein id: At1g05320.1 [Arabidopsis thaliana]
gi|25406838|pir||A86188 hypothetical protein [imported] -
Arabidopsis thaliana gi|2388564|gb|AAB71445.1| ESTs
gb|AA042402,gb|ATTS1380 come from this gene.
[Arabidopsis thaliana]
Length = 841
Score = 75.9 bits (185), Expect = 7e-13
Identities = 57/161 (35%), Positives = 83/161 (51%), Gaps = 13/161 (8%)
Frame = -3
Query: 795 ELEASLKNSLEELEIKKNEITLLQKQVTDLEQKLQLTSDKLSV-------------KGEV 655
ELEA+LK S EEL+ KK+ I L+ ++ +LEQK++L K V +
Sbjct: 685 ELEATLKKSQEELDAKKSVIVHLESKLNELEQKVKLADAKSKVSHIKHNHIFKPNLQETE 744
Query: 654 GVDQKDGLEVKSRDIGSSISSPSKRKSKKKSEATTAPTPSSSSETHVQTSQASPVINFKF 475
+++ +EVKSRD S S+P + K KK +A SSS +Q ++ ++ K
Sbjct: 745 STGKEEEVEVKSRDSDLSFSNPKQTKIKKNLDAA-----SSSGHVMIQKAETWHLMTLKI 799
Query: 474 ILGVALVSIIFGIILGKRY*YQQHDFLVLLFIMFGFSILFL 352
LGVALVS+I GII+ D V+L I +LFL
Sbjct: 800 ALGVALVSVILGIII------CLFDCFVVLRIFASTILLFL 834
>pir||F84730 probable myosin heavy chain [imported] - Arabidopsis thaliana
Length = 1269
Score = 73.6 bits (179), Expect = 3e-12
Identities = 51/153 (33%), Positives = 77/153 (49%), Gaps = 30/153 (19%)
Frame = -3
Query: 786 ASLKNSLEELEIKKNEITLLQKQVTDLEQKLQLTSDKLSVKGEVGVDQKDGLE------- 628
A L + L+E E E +L +QV L+++LQ + + + + ++ L
Sbjct: 1117 AELTSKLQEHEHIAGERDVLNEQVLQLQKELQAAQSSIDEQKRLKLRKRLSLNLNQWSKI 1176
Query: 627 ----------------------VKSRDIGSSISSPSKRKSKKKSEATTAPTPSSSS-ETH 517
VKSRDI S SSP+KRKSKKK EA+ + + SS + T
Sbjct: 1177 LNRKCSSQMLKLRLETEAMDVGVKSRDIDLSFSSPTKRKSKKKPEASLSSSSSSGNVTTP 1236
Query: 516 VQTSQASPVINFKFILGVALVSIIFGIILGKRY 418
QT+ S ++ K + GVAL+S+I GIILG++Y
Sbjct: 1237 TQTASTSHLMTVKIVTGVALISVIIGIILGRKY 1269
>ref|NP_341918.1| Hypothetical protein [Sulfolobus solfataricus]
gi|25392501|pir||E90181 hypothetical protein SSO0379
[imported] - Sulfolobus solfataricus
gi|13813526|gb|AAK40708.1| Hypothetical protein
[Sulfolobus solfataricus]
Length = 496
Score = 41.2 bits (95), Expect = 0.018
Identities = 26/58 (44%), Positives = 34/58 (57%)
Frame = -3
Query: 606 SSISSPSKRKSKKKSEATTAPTPSSSSETHVQTSQASPVINFKFILGVALVSIIFGII 433
SSI+S S S S +TT PT +SSS T V TS S + I+G+A+V II I+
Sbjct: 435 SSITSSSTTSSTTSSTSTTTPTSTSSSTTSVTTSGVSTSV----IIGIAIVVIIIVIV 488
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 706,339,002
Number of Sequences: 1393205
Number of extensions: 16648221
Number of successful extensions: 80067
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 61773
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 76004
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40896270532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)