Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000081A_C01 KMC000081A_c01
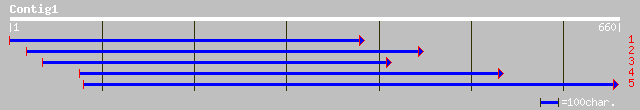
(660 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL58182.1|AC027037_4 putative far-red impaired response prot... 105 5e-22
dbj|BAB84393.1| P0529E05.9 [Oryza sativa (japonica cultivar-group)] 101 1e-20
gb|AAM15785.1|AC104428_6 Putative far-red impaired response prot... 99 7e-20
emb|CAD40515.1| OSJNBa0050F15.15 [Oryza sativa (japonica cultiva... 93 4e-18
gb|AAL82674.1|AC092387_22 putative transposase [Oryza sativa (ja... 93 4e-18
>gb|AAL58182.1|AC027037_4 putative far-red impaired response protein [Oryza sativa]
Length = 721
Score = 105 bits (262), Expect = 5e-22
Identities = 47/114 (41%), Positives = 71/114 (62%)
Frame = -2
Query: 641 GVRACHIMALMLGPKGGHESLGFTKTDLSNHIAKPKRERIQNGDAAAALSYLEGKADNDP 462
G+R C IM +M GG++ +GF DL N A+ K++RI+ DA +++L + + DP
Sbjct: 150 GLRTCQIMEVMENNNGGYDKVGFVTRDLYNFFARYKKKRIEGRDADLVVNHLMAQQEQDP 209
Query: 461 MFFYKFTKTGDESLENLFWCDGVSRMDYNVFGDVIAFDSTYKKNKYNKPLVVFL 300
FF++++ L NLFW D S++DY F DV+ FDSTY+ N+YN P V F+
Sbjct: 210 DFFFRYSIDEKGRLRNLFWADSQSQLDYEAFSDVVIFDSTYRVNRYNLPFVPFV 263
>dbj|BAB84393.1| P0529E05.9 [Oryza sativa (japonica cultivar-group)]
Length = 773
Score = 101 bits (251), Expect = 1e-20
Identities = 49/122 (40%), Positives = 70/122 (57%)
Frame = -2
Query: 659 DGMNLYGVRACHIMALMLGPKGGHESLGFTKTDLSNHIAKPKRERIQNGDAAAALSYLEG 480
D +N GVR+ +M GG +SL FT+ D N++ + + IQ GD A L YL+
Sbjct: 274 DILNDSGVRSKEGHEVMSRQAGGRQSLTFTRKDYKNYLRSKRMKSIQEGDTGAILQYLQD 333
Query: 479 KADNDPMFFYKFTKTGDESLENLFWCDGVSRMDYNVFGDVIAFDSTYKKNKYNKPLVVFL 300
K +P FFY DE + N+FW D S +D++ FGDVI FD+TY+ N Y +P +F+
Sbjct: 334 KQMENPSFFYAIQVDEDEMMTNIFWADARSVLDFDYFGDVICFDTTYRTNNYGRPFALFV 393
Query: 299 HV 294
V
Sbjct: 394 GV 395
>gb|AAM15785.1|AC104428_6 Putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)]
Length = 611
Score = 98.6 bits (244), Expect = 7e-20
Identities = 46/116 (39%), Positives = 67/116 (57%)
Frame = -2
Query: 641 GVRACHIMALMLGPKGGHESLGFTKTDLSNHIAKPKRERIQNGDAAAALSYLEGKADNDP 462
G+ +M ++ GG E GF DL N K K++RI GDA + Y++ + +D
Sbjct: 150 GLHQHQVMDVIERDHGGFEGAGFVTRDLYNFFVKMKKKRIDGGDADRVIKYMQARQKDDM 209
Query: 461 MFFYKFTKTGDESLENLFWCDGVSRMDYNVFGDVIAFDSTYKKNKYNKPLVVFLHV 294
F+Y++ L+ LFW D SR+DY+ FGDV+ FDSTY+ NKYN P + F+ V
Sbjct: 210 DFYYEYETDEAGCLKRLFWADPQSRIDYDAFGDVVVFDSTYRVNKYNLPFIPFVGV 265
>emb|CAD40515.1| OSJNBa0050F15.15 [Oryza sativa (japonica cultivar-group)]
Length = 1134
Score = 92.8 bits (229), Expect = 4e-18
Identities = 45/120 (37%), Positives = 65/120 (53%)
Frame = -2
Query: 653 MNLYGVRACHIMALMLGPKGGHESLGFTKTDLSNHIAKPKRERIQNGDAAAALSYLEGKA 474
+ + G+R +M +M + +GF DL N + + I+ DA L YL K
Sbjct: 590 LRMSGLRPFQVMEVMENNHDELDEVGFVMKDLYNFFTRYDMKNIKGHDAEDVLKYLTRKQ 649
Query: 473 DNDPMFFYKFTKTGDESLENLFWCDGVSRMDYNVFGDVIAFDSTYKKNKYNKPLVVFLHV 294
+ D FF+K+T + L N+FW D SR+DY FG V+ FDSTY+ NKYN P + F+ V
Sbjct: 650 EEDAEFFFKYTTDEEGRLRNVFWADAESRLDYAAFGGVVIFDSTYRVNKYNLPFIPFIGV 709
>gb|AAL82674.1|AC092387_22 putative transposase [Oryza sativa (japonica cultivar-group)]
Length = 311
Score = 92.8 bits (229), Expect = 4e-18
Identities = 46/121 (38%), Positives = 65/121 (53%)
Frame = -2
Query: 653 MNLYGVRACHIMALMLGPKGGHESLGFTKTDLSNHIAKPKRERIQNGDAAAALSYLEGKA 474
+ + G+R IM +M + +GF DL N + K + I+ DA YL K
Sbjct: 189 LRMSGLRPFQIMEVMENNHDELDEVGFVMKDLYNFFTQYKMKNIKGRDAEDVFKYLTKKQ 248
Query: 473 DNDPMFFYKFTKTGDESLENLFWCDGVSRMDYNVFGDVIAFDSTYKKNKYNKPLVVFLHV 294
+ D FF+K+T + L N+FW D SR+DY FG ++ FDSTY NKYN P + F+ V
Sbjct: 249 EKDAEFFFKYTTDEEWHLRNVFWADAESRIDYAAFGGIVIFDSTYCANKYNLPFIPFIGV 308
Query: 293 K 291
K
Sbjct: 309 K 309
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 563,520,494
Number of Sequences: 1393205
Number of extensions: 11863128
Number of successful extensions: 25504
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 23792
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25412
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28289785200
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)