Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC003452A_C01 KCC003452A_c01
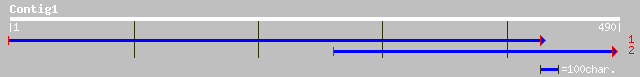
(490 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
prf||1209265Y chorion protein B7 41 0.009
prf||1209265R chorion protein B13 40 0.020
emb|CAA86030.1| LTBP-2 precursor [Homo sapiens] 38 0.058
ref|NP_000419.1| latent transforming growth factor beta binding ... 38 0.058
pir||A55494 latent transforming growth factor-beta-binding prote... 38 0.058
>prf||1209265Y chorion protein B7
Length = 161
Score = 40.8 bits (94), Expect = 0.009
Identities = 27/92 (29%), Positives = 38/92 (40%), Gaps = 11/92 (11%)
Frame = +3
Query: 219 CGSNMCD----------PSGMCASKQIKRAWDACPNGFTQVSKTNMTDLTSCEVSNSCGW 368
CGS CD P+G+ + + D C G +V D+ S+ CG+
Sbjct: 41 CGSGCCDRFCVCSNSAAPTGLSICSENRYKGDVCVCG--EVPFLGTADVCGDMCSSGCGY 98
Query: 369 -SRSCPDGSVYVSATCGDCWGCMEAGAGALAG 461
C DG V ++ +CG C GC G G G
Sbjct: 99 IDYGCGDGCVGITQSCGGCGGC---GCGCCGG 127
>prf||1209265R chorion protein B13
Length = 182
Score = 39.7 bits (91), Expect = 0.020
Identities = 26/88 (29%), Positives = 36/88 (40%), Gaps = 11/88 (12%)
Frame = +3
Query: 219 CGSNMCD----------PSGMCASKQIKRAWDACPNGFTQVSKTNMTDLTSCEVSNSCGW 368
CGS CD P+G+ + + D C G +V D+ S+ CG
Sbjct: 41 CGSRCCDRFCVCSNSAAPTGLSICSENRYNGDVCVCG--EVPFLGTADVCGDMCSSGCGC 98
Query: 369 -SRSCPDGSVYVSATCGDCWGCMEAGAG 449
C DG V ++ +CG C GC G G
Sbjct: 99 IDYGCGDGCVGITQSCGGCGGCGCGGCG 126
>emb|CAA86030.1| LTBP-2 precursor [Homo sapiens]
Length = 1821
Score = 38.1 bits (87), Expect = 0.058
Identities = 23/73 (31%), Positives = 33/73 (44%), Gaps = 3/73 (4%)
Frame = +3
Query: 219 CGSNMCDPSGMCASKQIKRAWDACPNGFTQVSKTNMTDLTSCEVSNSCGWSRSCPDG--- 389
C S P+G+C + + A AC NG+ D T+CE + C + CP G
Sbjct: 1055 CASRASCPTGLCLNTEGSFACSACENGYWV-----NEDGTACEDLDECAFPGVCPSGVCT 1109
Query: 390 SVYVSATCGDCWG 428
+ S +C DC G
Sbjct: 1110 NTAGSFSCKDCDG 1122
Score = 33.1 bits (74), Expect = 1.9
Identities = 25/92 (27%), Positives = 34/92 (36%), Gaps = 5/92 (5%)
Frame = +3
Query: 189 TLTVWTAHKACGS-NMCDPSGMCASKQIKRAWDA----CPNGFTQVSKTNMTDLTSCEVS 353
TL A + C N C+ G+C+ Q + C G+ V K + D+
Sbjct: 918 TLATSGATQECQDINECEQPGVCSGGQCTNTEGSYHCECDQGYIMVRKGHCQDI------ 971
Query: 354 NSCGWSRSCPDGSVYVSATCGDCWGCMEAGAG 449
N C +CPDG S C C E G
Sbjct: 972 NECRHPGTCPDGRCVNSPGSYTCLACEEGYRG 1003
>ref|NP_000419.1| latent transforming growth factor beta binding protein 2 [Homo
sapiens] gi|1699300|gb|AAB37459.1| latent transforming
growth factor-beta-binding protein-2; LTBP-2 [Homo
sapiens]
Length = 1821
Score = 38.1 bits (87), Expect = 0.058
Identities = 23/73 (31%), Positives = 33/73 (44%), Gaps = 3/73 (4%)
Frame = +3
Query: 219 CGSNMCDPSGMCASKQIKRAWDACPNGFTQVSKTNMTDLTSCEVSNSCGWSRSCPDG--- 389
C S P+G+C + + A AC NG+ D T+CE + C + CP G
Sbjct: 1055 CASRASCPTGLCLNTEGSFACSACENGYWV-----NEDGTACEDLDECAFPGVCPSGVCT 1109
Query: 390 SVYVSATCGDCWG 428
+ S +C DC G
Sbjct: 1110 NTAGSFSCKDCDG 1122
Score = 33.1 bits (74), Expect = 1.9
Identities = 25/92 (27%), Positives = 34/92 (36%), Gaps = 5/92 (5%)
Frame = +3
Query: 189 TLTVWTAHKACGS-NMCDPSGMCASKQIKRAWDA----CPNGFTQVSKTNMTDLTSCEVS 353
TL A + C N C+ G+C+ Q + C G+ V K + D+
Sbjct: 918 TLATSGATQECQDINECEQPGVCSGGQCTNTEGSYHCECDQGYIMVRKGHCQDI------ 971
Query: 354 NSCGWSRSCPDGSVYVSATCGDCWGCMEAGAG 449
N C +CPDG S C C E G
Sbjct: 972 NECRHPGTCPDGRCVNSPGSYTCLACEEGYRG 1003
>pir||A55494 latent transforming growth factor-beta-binding protein - human
Length = 1820
Score = 38.1 bits (87), Expect = 0.058
Identities = 23/73 (31%), Positives = 33/73 (44%), Gaps = 3/73 (4%)
Frame = +3
Query: 219 CGSNMCDPSGMCASKQIKRAWDACPNGFTQVSKTNMTDLTSCEVSNSCGWSRSCPDG--- 389
C S P+G+C + + A AC NG+ D T+CE + C + CP G
Sbjct: 1055 CASRASCPTGLCLNTEGSFACSACENGYWV-----NEDGTACEDLDECAFPGVCPSGVCT 1109
Query: 390 SVYVSATCGDCWG 428
+ S +C DC G
Sbjct: 1110 NTAGSFSCKDCDG 1122
Score = 33.1 bits (74), Expect = 1.9
Identities = 25/92 (27%), Positives = 34/92 (36%), Gaps = 5/92 (5%)
Frame = +3
Query: 189 TLTVWTAHKACGS-NMCDPSGMCASKQIKRAWDA----CPNGFTQVSKTNMTDLTSCEVS 353
TL A + C N C+ G+C+ Q + C G+ V K + D+
Sbjct: 918 TLATSGATQECQDINECEQPGVCSGGQCTNTEGSYHCECDQGYIMVRKGHCQDI------ 971
Query: 354 NSCGWSRSCPDGSVYVSATCGDCWGCMEAGAG 449
N C +CPDG S C C E G
Sbjct: 972 NECRHPGTCPDGRCVNSPGSYTCLACEEGYRG 1003