Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
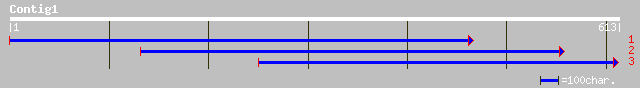
Query= KCC003374A_C01 KCC003374A_c01
(506 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T07927 protein disulfide-isomerase (EC 5.3.4.1) RB60 - Chla... 215 3e-55
gb|AAD55566.1|AF110784_1 protein disulfide isomerase precursor [... 155 2e-37
gb|AAO26314.1| protein disulphide isomerase [Elaeis guineensis] 94 1e-18
ref|NP_191056.2| protein disulfide isomerase family [Arabidopsis... 92 3e-18
pir||T06724 protein disulfide-isomerase homolog F28P10.60 - Arab... 92 3e-18
>pir||T07927 protein disulfide-isomerase (EC 5.3.4.1) RB60 - Chlamydomonas
reinhardtii gi|2708314|gb|AAC49896.1| protein disulfide
isomerase RB60 [Chlamydomonas reinhardtii]
gi|4104541|gb|AAD02069.1| protein disulfide isomerase
[Chlamydomonas reinhardtii]
Length = 532
Score = 215 bits (547), Expect = 3e-55
Identities = 105/106 (99%), Positives = 106/106 (99%)
Frame = +2
Query: 38 RKLEPIYKKLAKRFKKVDSVIIAKMDGTENEHPEIEVKGFPTILFYPAGSDRTPIVFEGG 217
+KLEPIYKKLAKRFKKVDSVIIAKMDGTENEHPEIEVKGFPTILFYPAGSDRTPIVFEGG
Sbjct: 427 KKLEPIYKKLAKRFKKVDSVIIAKMDGTENEHPEIEVKGFPTILFYPAGSDRTPIVFEGG 486
Query: 218 DRSLKSLTKFIKTNAKIPYELPKKGSDGDEGTSDDKDKPASDKDEL 355
DRSLKSLTKFIKTNAKIPYELPKKGSDGDEGTSDDKDKPASDKDEL
Sbjct: 487 DRSLKSLTKFIKTNAKIPYELPKKGSDGDEGTSDDKDKPASDKDEL 532
>gb|AAD55566.1|AF110784_1 protein disulfide isomerase precursor [Volvox carteri f.
nagariensis]
Length = 524
Score = 155 bits (393), Expect = 2e-37
Identities = 77/106 (72%), Positives = 89/106 (83%)
Frame = +2
Query: 38 RKLEPIYKKLAKRFKKVDSVIIAKMDGTENEHPEIEVKGFPTILFYPAGSDRTPIVFEGG 217
+KL+PIYKKLAKRFKKV SV+IAKMDGTENEHP ++VKGFPT++F+PAG D TPI FEGG
Sbjct: 423 KKLDPIYKKLAKRFKKVSSVVIAKMDGTENEHPLVDVKGFPTLIFFPAGEDATPIPFEGG 482
Query: 218 DRSLKSLTKFIKTNAKIPYELPKKGSDGDEGTSDDKDKPASDKDEL 355
DR+LKSLTKFIK NAK+PYELPKK S +E +DD S KDEL
Sbjct: 483 DRTLKSLTKFIKANAKVPYELPKK-SSAEESAADD---APSAKDEL 524
>gb|AAO26314.1| protein disulphide isomerase [Elaeis guineensis]
Length = 447
Score = 94.0 bits (232), Expect = 1e-18
Identities = 45/101 (44%), Positives = 64/101 (62%)
Frame = +2
Query: 44 LEPIYKKLAKRFKKVDSVIIAKMDGTENEHPEIEVKGFPTILFYPAGSDRTPIVFEGGDR 223
LEP Y KLAK + ++S++IAKMDGT NEHP +V GFPT+LF+PAG+ V DR
Sbjct: 337 LEPTYNKLAKHLRGIESLVIAKMDGTSNEHPRAKVDGFPTLLFFPAGNKSFDPVTVDTDR 396
Query: 224 SLKSLTKFIKTNAKIPYELPKKGSDGDEGTSDDKDKPASDK 346
++ + KFIK +A IP++L + S ++ D P +K
Sbjct: 397 TVVAFYKFIKKHAAIPFKLQRPASAAKTDSATDGSAPVGEK 437
Score = 32.0 bits (71), Expect = 4.5
Identities = 15/34 (44%), Positives = 22/34 (64%), Gaps = 2/34 (5%)
Frame = +2
Query: 89 DSVIIAKMDGTENEH--PEIEVKGFPTILFYPAG 184
+ V +AK+D TE + EV+GFPT+LF+ G
Sbjct: 12 EDVALAKVDATEENELAQKYEVQGFPTVLFFVDG 45
>ref|NP_191056.2| protein disulfide isomerase family [Arabidopsis thaliana]
gi|18072841|emb|CAC81067.1| ERp72 [Arabidopsis thaliana]
gi|20260432|gb|AAM13114.1| protein
disulfide-isomerase-like protein [Arabidopsis thaliana]
gi|23197928|gb|AAN15491.1| protein
disulfide-isomerase-like protein [Arabidopsis thaliana]
Length = 579
Score = 92.4 bits (228), Expect = 3e-18
Identities = 48/109 (44%), Positives = 66/109 (60%), Gaps = 3/109 (2%)
Frame = +2
Query: 38 RKLEPIYKKLAKRFKKVDSVIIAKMDGTENEHPEIEVKGFPTILFYPAGSDRTPIVFEGG 217
+ EPIY KL K K +DS+++AKMDGT NEHP + GFPTILF+P G+ +
Sbjct: 471 QSFEPIYNKLGKYLKGIDSLVVAKMDGTSNEHPRAKADGFPTILFFPGGNKSFDPIAVDV 530
Query: 218 DRSLKSLTKFIKTNAKIPYELPKKGSDG---DEGTSDDKDKPASDKDEL 355
DR++ L KF+K +A IP++L K + SD+K + S KDEL
Sbjct: 531 DRTVVELYKFLKKHASIPFKLEKPATPEPVISTMKSDEKIEGDSSKDEL 579
>pir||T06724 protein disulfide-isomerase homolog F28P10.60 - Arabidopsis
thaliana gi|4678297|emb|CAB41088.1| protein
disulfide-isomerase-like protein [Arabidopsis thaliana]
Length = 566
Score = 92.4 bits (228), Expect = 3e-18
Identities = 48/109 (44%), Positives = 66/109 (60%), Gaps = 3/109 (2%)
Frame = +2
Query: 38 RKLEPIYKKLAKRFKKVDSVIIAKMDGTENEHPEIEVKGFPTILFYPAGSDRTPIVFEGG 217
+ EPIY KL K K +DS+++AKMDGT NEHP + GFPTILF+P G+ +
Sbjct: 458 QSFEPIYNKLGKYLKGIDSLVVAKMDGTSNEHPRAKADGFPTILFFPGGNKSFDPIAVDV 517
Query: 218 DRSLKSLTKFIKTNAKIPYELPKKGSDG---DEGTSDDKDKPASDKDEL 355
DR++ L KF+K +A IP++L K + SD+K + S KDEL
Sbjct: 518 DRTVVELYKFLKKHASIPFKLEKPATPEPVISTMKSDEKIEGDSSKDEL 566