Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC003239A_C01 KCC003239A_c01
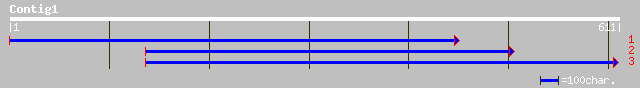
(611 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_172136.1| callose synthase (1,3-beta-glucan synthase) fam... 97 1e-19
pir||F86200 protein F12K11.17 [imported] - Arabidopsis thaliana ... 97 1e-19
dbj|BAB89139.1| OJ1029_F04.4 [Oryza sativa (japonica cultivar-gr... 91 2e-17
dbj|BAB02389.1| glucan synthase-like protein [Arabidopsis thaliana] 88 8e-17
ref|NP_188075.1| callose synthase (1,3-beta-glucan synthase) fam... 86 4e-16
>ref|NP_172136.1| callose synthase (1,3-beta-glucan synthase) family [Arabidopsis
thaliana]
Length = 1933
Score = 97.4 bits (241), Expect = 1e-19
Identities = 72/219 (32%), Positives = 108/219 (48%), Gaps = 16/219 (7%)
Frame = +3
Query: 3 LTPLYEEDVMYALDSKALAKETGLKMRKMTDLLGETDDSISLMSYLKAMFPQEWSNFKER 182
LTP Y+EDV+Y+ + +L E +D I+++ YL+ ++P+EWSN+ ER
Sbjct: 1086 LTPYYKEDVLYSEE----------------ELNKENEDGITILFYLQRIYPEEWSNYCER 1129
Query: 183 MKTLNPDINVKDLSEHDFAPGCDMYEFKLELQMWASLRGQLLARTVHGMMLNEKAL---- 350
+ L +++ KD +E +L+ W S RGQ L+RTV GMM AL
Sbjct: 1130 VNDLKRNLSEKDKAE--------------QLRQWVSYRGQTLSRTVRGMMYYRVALELQC 1175
Query: 351 -----DELARLENPQPPNMTELEYKRY---IHQLTSCKFEYVVTPQTYGKNRLSKDLRLK 506
+E A P E + K + L KF YVV+ Q YG + S + R +
Sbjct: 1176 FQEYTEENATNGGYLPSESNEDDRKAFSDRARALADLKFTYVVSCQVYGNQKKSSESRDR 1235
Query: 507 WLASSIDILMGKYPRLKVAFLDNADSD-NGPAQ---YSV 611
++I LM KYP L+VA++D + NG +Q YSV
Sbjct: 1236 SCYNNILQLMLKYPSLRVAYIDEREETVNGKSQKVFYSV 1274
>pir||F86200 protein F12K11.17 [imported] - Arabidopsis thaliana
gi|6692688|gb|AAF24822.1|AC007592_15 F12K11.17
[Arabidopsis thaliana]
Length = 1930
Score = 97.4 bits (241), Expect = 1e-19
Identities = 72/219 (32%), Positives = 108/219 (48%), Gaps = 16/219 (7%)
Frame = +3
Query: 3 LTPLYEEDVMYALDSKALAKETGLKMRKMTDLLGETDDSISLMSYLKAMFPQEWSNFKER 182
LTP Y+EDV+Y+ + +L E +D I+++ YL+ ++P+EWSN+ ER
Sbjct: 1083 LTPYYKEDVLYSEE----------------ELNKENEDGITILFYLQRIYPEEWSNYCER 1126
Query: 183 MKTLNPDINVKDLSEHDFAPGCDMYEFKLELQMWASLRGQLLARTVHGMMLNEKAL---- 350
+ L +++ KD +E +L+ W S RGQ L+RTV GMM AL
Sbjct: 1127 VNDLKRNLSEKDKAE--------------QLRQWVSYRGQTLSRTVRGMMYYRVALELQC 1172
Query: 351 -----DELARLENPQPPNMTELEYKRY---IHQLTSCKFEYVVTPQTYGKNRLSKDLRLK 506
+E A P E + K + L KF YVV+ Q YG + S + R +
Sbjct: 1173 FQEYTEENATNGGYLPSESNEDDRKAFSDRARALADLKFTYVVSCQVYGNQKKSSESRDR 1232
Query: 507 WLASSIDILMGKYPRLKVAFLDNADSD-NGPAQ---YSV 611
++I LM KYP L+VA++D + NG +Q YSV
Sbjct: 1233 SCYNNILQLMLKYPSLRVAYIDEREETVNGKSQKVFYSV 1271
>dbj|BAB89139.1| OJ1029_F04.4 [Oryza sativa (japonica cultivar-group)]
Length = 1877
Score = 90.5 bits (223), Expect = 2e-17
Identities = 61/193 (31%), Positives = 93/193 (47%)
Frame = +3
Query: 3 LTPLYEEDVMYALDSKALAKETGLKMRKMTDLLGETDDSISLMSYLKAMFPQEWSNFKER 182
LTP Y E+V+Y+ +L + +D IS++ YL+ ++P EW NF ER
Sbjct: 1038 LTPYYNEEVLYSSH----------------ELNKKNEDGISILFYLQKIYPDEWKNFLER 1081
Query: 183 MKTLNPDINVKDLSEHDFAPGCDMYEFKLELQMWASLRGQLLARTVHGMMLNEKALDELA 362
+ D + G + ++++WAS RGQ LARTV GMM +AL+
Sbjct: 1082 IGV--------DPENEEAVKG-----YMDDVRIWASYRGQTLARTVRGMMYYRRALELQC 1128
Query: 363 RLENPQPPNMTELEYKRYIHQLTSCKFEYVVTPQTYGKNRLSKDLRLKWLASSIDILMGK 542
+ + E + KF YVV+ Q YG ++ SKD R K L +I LM
Sbjct: 1129 YEDMTNAQADLDGEESARSKAIADIKFTYVVSCQLYGMHKASKDSREKGLYENILNLMLT 1188
Query: 543 YPRLKVAFLDNAD 581
YP L++A++D +
Sbjct: 1189 YPALRIAYIDEKE 1201
>dbj|BAB02389.1| glucan synthase-like protein [Arabidopsis thaliana]
Length = 1972
Score = 88.2 bits (217), Expect = 8e-17
Identities = 66/198 (33%), Positives = 92/198 (46%), Gaps = 5/198 (2%)
Frame = +3
Query: 3 LTPLYEEDVMYALDSKALAKETGLKMRKMTDLLGETDDSISLMSYLKAMFPQEWSNFKER 182
LTP Y+ED+ Y+ T+ L T S+S++ Y++ +FP EW NF ER
Sbjct: 1143 LTPHYQEDINYS-----------------TNELHSTKSSVSIIFYMQKIFPDEWKNFLER 1185
Query: 183 MKTLNPDINVKDLSEHDFAPGCDMYEFKLELQMWASLRGQLLARTVHGMMLNEKALDELA 362
M N D K+ E EL+ WAS RGQ L+RTV GMM +AL A
Sbjct: 1186 MGCDNLDALKKEGKEE-------------ELRNWASFRGQTLSRTVRGMMYCREALKLQA 1232
Query: 363 RLENPQPPNMTELEYKR-----YIHQLTSCKFEYVVTPQTYGKNRLSKDLRLKWLASSID 527
L+ ++E + L KF YVV+ Q +G + S D A I
Sbjct: 1233 FLDMADDEGYKDVERSNRPLAAQLDALADMKFTYVVSCQMFGAQKSSGDPH----AQDIL 1288
Query: 528 ILMGKYPRLKVAFLDNAD 581
LM KYP L+VA+++ +
Sbjct: 1289 DLMIKYPSLRVAYVEERE 1306
>ref|NP_188075.1| callose synthase (1,3-beta-glucan synthase) family [Arabidopsis
thaliana]
Length = 1973
Score = 85.9 bits (211), Expect = 4e-16
Identities = 66/202 (32%), Positives = 92/202 (44%), Gaps = 9/202 (4%)
Frame = +3
Query: 3 LTPLYEEDVMYALDSKALAKETGLKMRKMTDLLGETDDSISLMSYLKAMFPQEWSNFKER 182
LTP Y+ED+ Y+ T+ L T S+S++ Y++ +FP EW NF ER
Sbjct: 1140 LTPHYQEDINYS-----------------TNELHSTKSSVSIIFYMQKIFPDEWKNFLER 1182
Query: 183 MKTLNPDINVKDLSEHDFAPGCDMYEFKLELQMWASLRGQLLARTVHGMMLNEKAL---- 350
M N D K+ E EL+ WAS RGQ L+RTV GMM +AL
Sbjct: 1183 MGCDNLDALKKEGKEE-------------ELRNWASFRGQTLSRTVRGMMYCREALKLQA 1229
Query: 351 -----DELARLENPQPPNMTELEYKRYIHQLTSCKFEYVVTPQTYGKNRLSKDLRLKWLA 515
D+ LE + + + L KF YVV+ Q +G + S D A
Sbjct: 1230 FLDMADDEDILEGYKDVERSNRPLAAQLDALADMKFTYVVSCQMFGAQKSSGDPH----A 1285
Query: 516 SSIDILMGKYPRLKVAFLDNAD 581
I LM KYP L+VA+++ +
Sbjct: 1286 QDILDLMIKYPSLRVAYVEERE 1307