Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC003096A_C01 KCC003096A_c01
(887 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
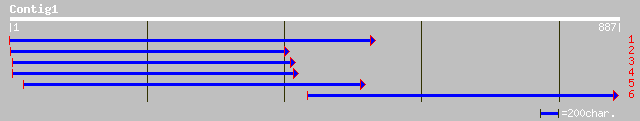
pir||T52098 probable nuclear transport factor importin alpha [im... 340 2e-92
gb|AAK32824.1|AF361811_1 AT3g06720/F3E22_14 [Arabidopsis thalian... 340 2e-92
ref|NP_567485.1| expressed protein [Arabidopsis thaliana] 340 2e-92
gb|AAK38726.1|AF369706_1 importin alpha 1 [Capsicum annuum] 339 3e-92
dbj|BAA31165.1| NLS receptor [Oryza sativa] gi|3273245|dbj|BAA31... 336 4e-91
>pir||T52098 probable nuclear transport factor importin alpha [imported] -
Arabidopsis thaliana gi|2950210|emb|CAA74965.1| Importin
alpha-like protein [Arabidopsis thaliana]
Length = 535
Score = 340 bits (872), Expect = 2e-92
Identities = 172/268 (64%), Positives = 205/268 (76%), Gaps = 3/268 (1%)
Frame = +2
Query: 92 LRPDAS---RKKEYKKGIDAEEARRKREDNIIALRQNKRDENLQKKRSTFAPASANANFG 262
LRP+A R+ YK +DAEE RR+REDN++ +R++KR+E+LQKKR A+ F
Sbjct: 3 LRPNAKTEVRRNRYKVAVDAEEGRRRREDNMVEIRKSKREESLQKKRREGLQANQLPQFA 62
Query: 263 IEDSTKHAVGRQQLDELPMMVHGVFNGTVEQQYDCTQRFRKLLSIERNPPIEEVIKTGVI 442
+ ++L+ LP MV GV++ Q + T +FRKLLSIER+PPIEEVI GV+
Sbjct: 63 PSSVPASSTVEKKLESLPAMVGGVWSDDRSLQLEATTQFRKLLSIERSPPIEEVIDAGVV 122
Query: 443 PKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNAVPIFVQLLASPNDDVREQ 622
P+FVEFL R D PQLQFEAAWALTN+ASGTSE+TKVVI+H AVPIFVQLLAS +DDVREQ
Sbjct: 123 PRFVEFLTREDYPQLQFEAAWALTNIASGTSENTKVVIEHGAVPIFVQLLASQSDDVREQ 182
Query: 623 AVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRNATWTLSNFCRGKPAPNF 802
AVWALGN+AGDSP+CRD VLG AL PLL QL E++K+SMLRNATWTLSNFCRGKP P F
Sbjct: 183 AVWALGNVAGDSPRCRDLVLGQGALIPLLSQLNEHAKLSMLRNATWTLSNFCRGKPQPPF 242
Query: 803 IVTRQALPTLARLIHHSDEEVLTDACWA 886
R ALP L RLIH +DEEVLTDACWA
Sbjct: 243 DQVRPALPALERLIHSTDEEVLTDACWA 270
Score = 60.1 bits (144), Expect = 5e-08
Identities = 45/162 (27%), Positives = 79/162 (47%), Gaps = 2/162 (1%)
Frame = +2
Query: 407 PPIEEVIKTGVIPKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNAVPIFVQ 586
PP ++V +P + D L +A WAL+ ++ GT++ + VI+ VP V+
Sbjct: 240 PPFDQV--RPALPALERLIHSTDEEVLT-DACWALSYLSDGTNDKVQSVIEAGVVPRLVE 296
Query: 587 LLASPNDDVREQAVWALGN-IAGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRNATWT 763
LL + V A+ ++GN + GD + + V+ A P LL L N K S+ + A WT
Sbjct: 297 LLQHQSPSVLIPALRSIGNNVTGDDLQTQ-CVIKSCAPPSLLGLLTHNHKKSIKKEACWT 355
Query: 764 LSNFCRG-KPAPNFIVTRQALPTLARLIHHSDEEVLTDACWA 886
+SN G + + + L L+ +++ ++ +A WA
Sbjct: 356 ISNITAGNRDQIQAVCEAGLICPLVNLLQNAEFDIKKEAAWA 397
Score = 59.3 bits (142), Expect = 8e-08
Identities = 43/164 (26%), Positives = 77/164 (46%), Gaps = 3/164 (1%)
Frame = +2
Query: 389 LSIERNPPIEEVIKTGVIPKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNA 568
LS N ++ VI+ GV+P+ VE LQ H +P + A ++ N +G T+ VI A
Sbjct: 274 LSDGTNDKVQSVIEAGVVPRLVELLQ-HQSPSVLIPALRSIGNNVTGDDLQTQCVIKSCA 332
Query: 569 VPIFVQLLA-SPNDDVREQAVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISML 745
P + LL + ++++A W + NI + V + PL+ L +N++ +
Sbjct: 333 PPSLLGLLTHNHKKSIKKEACWTISNITAGNRDQIQAVCEAGLICPLVNLL-QNAEFDIK 391
Query: 746 RNATWTLSNFCRGKPAP--NFIVTRQALPTLARLIHHSDEEVLT 871
+ A W +SN G ++V + + L L+ D ++T
Sbjct: 392 KEAAWAISNATSGGSPDQIKYMVEQGVVKPLCDLLVCPDPRIIT 435
>gb|AAK32824.1|AF361811_1 AT3g06720/F3E22_14 [Arabidopsis thaliana]
gi|16974501|gb|AAL31160.1| AT3g06720/F3E22_14
[Arabidopsis thaliana]
Length = 535
Score = 340 bits (872), Expect = 2e-92
Identities = 172/268 (64%), Positives = 205/268 (76%), Gaps = 3/268 (1%)
Frame = +2
Query: 92 LRPDAS---RKKEYKKGIDAEEARRKREDNIIALRQNKRDENLQKKRSTFAPASANANFG 262
LRP+A R+ YK +DAEE RR+REDN++ +R++KR+E+LQKKR A+ F
Sbjct: 3 LRPNAKTEVRRNRYKVAVDAEEGRRRREDNMVEIRKSKREESLQKKRREGLQANQLPQFA 62
Query: 263 IEDSTKHAVGRQQLDELPMMVHGVFNGTVEQQYDCTQRFRKLLSIERNPPIEEVIKTGVI 442
+ ++L+ LP MV GV++ Q + T +FRKLLSIER+PPIEEVI GV+
Sbjct: 63 PSSVPASSTVEKKLESLPAMVGGVWSDDRSLQLEATTQFRKLLSIERSPPIEEVIDAGVV 122
Query: 443 PKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNAVPIFVQLLASPNDDVREQ 622
P+FVEFL R D PQLQFEAAWALTN+ASGTSE+TKVVI+H AVPIFVQLLAS +DDVREQ
Sbjct: 123 PRFVEFLTREDYPQLQFEAAWALTNIASGTSENTKVVIEHGAVPIFVQLLASQSDDVREQ 182
Query: 623 AVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRNATWTLSNFCRGKPAPNF 802
AVWALGN+AGDSP+CRD VLG AL PLL QL E++K+SMLRNATWTLSNFCRGKP P F
Sbjct: 183 AVWALGNVAGDSPRCRDLVLGQGALIPLLSQLNEHAKLSMLRNATWTLSNFCRGKPQPPF 242
Query: 803 IVTRQALPTLARLIHHSDEEVLTDACWA 886
R ALP L RLIH +DEEVLTDACWA
Sbjct: 243 DQVRPALPALERLIHSTDEEVLTDACWA 270
Score = 66.6 bits (161), Expect = 5e-10
Identities = 45/161 (27%), Positives = 76/161 (46%), Gaps = 1/161 (0%)
Frame = +2
Query: 407 PPIEEVIKTGVIPKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNAVPIFVQ 586
PP ++V +P + D L +A WAL+ ++ GT++ + VI+ VP V+
Sbjct: 240 PPFDQV--RPALPALERLIHSTDEEVLT-DACWALSYLSDGTNDKIQSVIEAGVVPRLVE 296
Query: 587 LLASPNDDVREQAVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRNATWTL 766
LL + V A+ ++GNI V+ H AL LL L N K S+ + A WT+
Sbjct: 297 LLQHQSPSVLIPALRSIGNIVTGDDLQTQCVISHGALLSLLSLLTHNHKKSIKKEACWTI 356
Query: 767 SNFCRG-KPAPNFIVTRQALPTLARLIHHSDEEVLTDACWA 886
SN G + + + L L+ +++ ++ +A WA
Sbjct: 357 SNITAGNRDQIQAVCEAGLICPLVNLLQNAEFDIKKEAAWA 397
Score = 65.1 bits (157), Expect = 2e-09
Identities = 44/164 (26%), Positives = 79/164 (47%), Gaps = 3/164 (1%)
Frame = +2
Query: 389 LSIERNPPIEEVIKTGVIPKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNA 568
LS N I+ VI+ GV+P+ VE LQ H +P + A ++ N+ +G T+ VI H A
Sbjct: 274 LSDGTNDKIQSVIEAGVVPRLVELLQ-HQSPSVLIPALRSIGNIVTGDDLQTQCVISHGA 332
Query: 569 VPIFVQLLA-SPNDDVREQAVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISML 745
+ + LL + ++++A W + NI + V + PL+ L +N++ +
Sbjct: 333 LLSLLSLLTHNHKKSIKKEACWTISNITAGNRDQIQAVCEAGLICPLVNLL-QNAEFDIK 391
Query: 746 RNATWTLSNFCRGKPAP--NFIVTRQALPTLARLIHHSDEEVLT 871
+ A W +SN G ++V + + L L+ D ++T
Sbjct: 392 KEAAWAISNATSGGSPDQIKYMVEQGVVKPLCDLLVCPDPRIIT 435
Score = 37.4 bits (85), Expect = 0.34
Identities = 31/116 (26%), Positives = 53/116 (44%), Gaps = 3/116 (2%)
Frame = +2
Query: 413 IEEVIKTGVIPKFVEFLQRHDTPQLQFEAAWALTNVASGTS-EHTKVVIDHNAVPIFVQL 589
I+ V + G+I V LQ + ++ EAAWA++N SG S + K +++ V L
Sbjct: 367 IQAVCEAGLICPLVNLLQNAEF-DIKKEAAWAISNATSGGSPDQIKYMVEQGVVKPLCDL 425
Query: 590 LASPNDDVREQAVWALGNI--AGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRN 751
L P+ + + L NI G++ K N L++ + KI L++
Sbjct: 426 LVCPDPRIITVCLEGLENILKVGEAEKVTGNTGDVNFYAQLIDDAEGLEKIENLQS 481
>ref|NP_567485.1| expressed protein [Arabidopsis thaliana]
Length = 444
Score = 340 bits (871), Expect = 2e-92
Identities = 173/268 (64%), Positives = 206/268 (76%), Gaps = 3/268 (1%)
Frame = +2
Query: 92 LRPDAS---RKKEYKKGIDAEEARRKREDNIIALRQNKRDENLQKKRSTFAPASANANFG 262
LRP+A R+ YK +DAEE RR+REDN++ +R++KR+E+LQKKR A+ F
Sbjct: 3 LRPNAKTEVRRNRYKVAVDAEEGRRRREDNMVEIRKSKREESLQKKRREGLQANQLPQFA 62
Query: 263 IEDSTKHAVGRQQLDELPMMVHGVFNGTVEQQYDCTQRFRKLLSIERNPPIEEVIKTGVI 442
H + ++L+ LP MV GV++ Q + T +FRKLLSIER+PPIEEVI GV+
Sbjct: 63 PLLFLLHQLLEKKLESLPAMVGGVWSDDRSLQLEATTQFRKLLSIERSPPIEEVIDAGVV 122
Query: 443 PKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNAVPIFVQLLASPNDDVREQ 622
P+FVEFL R D PQLQFEAAWALTN+ASGTSE+TKVVI+H AVPIFVQLLAS +DDVREQ
Sbjct: 123 PRFVEFLTREDYPQLQFEAAWALTNIASGTSENTKVVIEHGAVPIFVQLLASQSDDVREQ 182
Query: 623 AVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRNATWTLSNFCRGKPAPNF 802
AVWALGN+AGDSP+CRD VLG AL PLL QL E++K+SMLRNATWTLSNFCRGKP P F
Sbjct: 183 AVWALGNVAGDSPRCRDLVLGQGALIPLLSQLNEHAKLSMLRNATWTLSNFCRGKPQPPF 242
Query: 803 IVTRQALPTLARLIHHSDEEVLTDACWA 886
R ALP L RLIH +DEEVLTDACWA
Sbjct: 243 DQVRPALPALERLIHSTDEEVLTDACWA 270
Score = 66.6 bits (161), Expect = 5e-10
Identities = 45/161 (27%), Positives = 76/161 (46%), Gaps = 1/161 (0%)
Frame = +2
Query: 407 PPIEEVIKTGVIPKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNAVPIFVQ 586
PP ++V +P + D L +A WAL+ ++ GT++ + VI+ VP V+
Sbjct: 240 PPFDQV--RPALPALERLIHSTDEEVLT-DACWALSYLSDGTNDKIQSVIEAGVVPRLVE 296
Query: 587 LLASPNDDVREQAVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRNATWTL 766
LL + V A+ ++GNI V+ H AL LL L N K S+ + A WT+
Sbjct: 297 LLQHQSPSVLIPALRSIGNIVTGDDLQTQCVISHGALLSLLSLLTHNHKKSIKKEACWTI 356
Query: 767 SNFCRG-KPAPNFIVTRQALPTLARLIHHSDEEVLTDACWA 886
SN G + + + L L+ +++ ++ +A WA
Sbjct: 357 SNITAGNRDQIQAVCEAGLICPLVNLLQNAEFDIKKEAAWA 397
Score = 63.9 bits (154), Expect = 3e-09
Identities = 39/133 (29%), Positives = 67/133 (50%), Gaps = 1/133 (0%)
Frame = +2
Query: 389 LSIERNPPIEEVIKTGVIPKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNA 568
LS N I+ VI+ GV+P+ VE LQ H +P + A ++ N+ +G T+ VI H A
Sbjct: 274 LSDGTNDKIQSVIEAGVVPRLVELLQ-HQSPSVLIPALRSIGNIVTGDDLQTQCVISHGA 332
Query: 569 VPIFVQLLA-SPNDDVREQAVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISML 745
+ + LL + ++++A W + NI + V + PL+ L +N++ +
Sbjct: 333 LLSLLSLLTHNHKKSIKKEACWTISNITAGNRDQIQAVCEAGLICPLVNLL-QNAEFDIK 391
Query: 746 RNATWTLSNFCRG 784
+ A W +SN G
Sbjct: 392 KEAAWAISNATSG 404
>gb|AAK38726.1|AF369706_1 importin alpha 1 [Capsicum annuum]
Length = 535
Score = 339 bits (870), Expect = 3e-92
Identities = 173/268 (64%), Positives = 204/268 (75%), Gaps = 3/268 (1%)
Frame = +2
Query: 92 LRPDAS---RKKEYKKGIDAEEARRKREDNIIALRQNKRDENLQKKRSTFAPASANANFG 262
LRP RKK YK G+DA+EARR+REDN++ +R+NKR++NL KKR +
Sbjct: 3 LRPSTRTEVRKKAYKIGVDADEARRRREDNLVEIRKNKREDNLLKKRREGLLFHSQQQLP 62
Query: 263 IEDSTKHAVGRQQLDELPMMVHGVFNGTVEQQYDCTQRFRKLLSIERNPPIEEVIKTGVI 442
T A+ ++L+ +P+MV GV++ Q + T FRKLLSIER+PPI+EVIK GVI
Sbjct: 63 DAAQTPDAI-EKRLESIPVMVQGVWSEDPAAQIEATTHFRKLLSIERSPPIDEVIKAGVI 121
Query: 443 PKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNAVPIFVQLLASPNDDVREQ 622
P+FVEFL RHD PQLQFEAAWALTNVASGTSEHT+VVIDH AVP+FVQLL+S +DDVREQ
Sbjct: 122 PRFVEFLGRHDLPQLQFEAAWALTNVASGTSEHTRVVIDHGAVPMFVQLLSSASDDVREQ 181
Query: 623 AVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRNATWTLSNFCRGKPAPNF 802
AVWALGN+AGDSP CRD VL AL PLL QL E+SK+SMLRNATWTLSNFCRGKP F
Sbjct: 182 AVWALGNVAGDSPSCRDLVLSQGALMPLLSQLNEHSKLSMLRNATWTLSNFCRGKPPTPF 241
Query: 803 IVTRQALPTLARLIHHSDEEVLTDACWA 886
+ ALP L +LIH SDEEVLTDACWA
Sbjct: 242 EAVKPALPVLQQLIHMSDEEVLTDACWA 269
Score = 70.5 bits (171), Expect = 4e-11
Identities = 44/161 (27%), Positives = 74/161 (45%), Gaps = 1/161 (0%)
Frame = +2
Query: 407 PPIEEVIKTGVIPKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNAVPIFVQ 586
PP +P + + D L +A WAL+ ++ G ++ + VID P V+
Sbjct: 237 PPTPFEAVKPALPVLQQLIHMSDEEVLT-DACWALSYLSDGPNDKIQAVIDAGVCPRLVE 295
Query: 587 LLASPNDDVREQAVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRNATWTL 766
LL P+ V A+ GNI YV+ + LP L + L +N K S+ + A WT+
Sbjct: 296 LLLHPSPTVLIPALRTAGNIVTGDDAQTQYVIDNRVLPCLYQLLTQNHKKSIKKEACWTI 355
Query: 767 SNFCRGKPAP-NFIVTRQALPTLARLIHHSDEEVLTDACWA 886
SN G A ++ + L L+ +++ ++ +A WA
Sbjct: 356 SNITAGNRAQIQAVIEANIILPLVHLLQNAEFDIKKEAAWA 396
Score = 65.5 bits (158), Expect = 1e-09
Identities = 45/164 (27%), Positives = 77/164 (46%), Gaps = 3/164 (1%)
Frame = +2
Query: 389 LSIERNPPIEEVIKTGVIPKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNA 568
LS N I+ VI GV P+ VE L H +P + A N+ +G T+ VID+
Sbjct: 273 LSDGPNDKIQAVIDAGVCPRLVELLL-HPSPTVLIPALRTAGNIVTGDDAQTQYVIDNRV 331
Query: 569 VPIFVQLLASPND-DVREQAVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISML 745
+P QLL + ++++A W + NI + V+ N + PL+ L +N++ +
Sbjct: 332 LPCLYQLLTQNHKKSIKKEACWTISNITAGNRAQIQAVIEANIILPLVHLL-QNAEFDIK 390
Query: 746 RNATWTLSNFCRG--KPAPNFIVTRQALPTLARLIHHSDEEVLT 871
+ A W +SN G F+ ++ + L L+ D ++T
Sbjct: 391 KEAAWAISNATSGGSNEQIRFLASQGCIKPLCDLLICPDPRIVT 434
>dbj|BAA31165.1| NLS receptor [Oryza sativa] gi|3273245|dbj|BAA31166.1| NLS receptor
[Oryza sativa (japonica cultivar-group)]
gi|6498466|dbj|BAA87855.1| unnamed protein product
[Oryza sativa (japonica cultivar-group)]
Length = 526
Score = 336 bits (861), Expect = 4e-91
Identities = 168/263 (63%), Positives = 207/263 (77%), Gaps = 4/263 (1%)
Frame = +2
Query: 110 RKKEYKKGIDAEEARRKREDNIIALRQNKRDENLQKKR----STFAPASANANFGIEDST 277
R+ YK +DAEE RR+REDN++ +R+++R+E+L KKR AP A+A G++
Sbjct: 12 RRNRYKVAVDAEEGRRRREDNMVEIRKSRREESLLKKRREGLQAQAPVPASAATGVD--- 68
Query: 278 KHAVGRQQLDELPMMVHGVFNGTVEQQYDCTQRFRKLLSIERNPPIEEVIKTGVIPKFVE 457
++L+ LP M+ GV++ Q + T +FRKLLSIER+PPIEEVI++GV+P+FV+
Sbjct: 69 ------KKLESLPAMIGGVYSDDNNLQLEATTQFRKLLSIERSPPIEEVIQSGVVPRFVQ 122
Query: 458 FLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNAVPIFVQLLASPNDDVREQAVWAL 637
FL R D PQLQFEAAWALTN+ASGTSE+TKVVIDH AVPIFV+LL S +DDVREQAVWAL
Sbjct: 123 FLTREDFPQLQFEAAWALTNIASGTSENTKVVIDHGAVPIFVKLLGSSSDDVREQAVWAL 182
Query: 638 GNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRNATWTLSNFCRGKPAPNFIVTRQ 817
GN+AGDSPKCRD VL + AL PLL QL E++K+SMLRNATWTLSNFCRGKP P+F TR
Sbjct: 183 GNVAGDSPKCRDLVLANGALLPLLAQLNEHTKLSMLRNATWTLSNFCRGKPQPSFEQTRP 242
Query: 818 ALPTLARLIHHSDEEVLTDACWA 886
ALP LARLIH +DEEVLTDACWA
Sbjct: 243 ALPALARLIHSNDEEVLTDACWA 265
Score = 72.8 bits (177), Expect = 7e-12
Identities = 43/150 (28%), Positives = 73/150 (48%), Gaps = 1/150 (0%)
Frame = +2
Query: 440 IPKFVEFLQRHDTPQLQFEAAWALTNVASGTSEHTKVVIDHNAVPIFVQLLASPNDDVRE 619
+P + +D L +A WAL+ ++ GT++ + VI+ P V+LL P+ V
Sbjct: 244 LPALARLIHSNDEEVLT-DACWALSYLSDGTNDKIQAVIEAGVCPRLVELLLHPSPSVLI 302
Query: 620 QAVWALGNIAGDSPKCRDYVLGHNALPPLLEQLKENSKISMLRNATWTLSNFCRG-KPAP 796
A+ +GNI ++ H ALP LL L +N K S+ + A WT+SN G K
Sbjct: 303 PALRTVGNIVTGDDAQTQCIIDHQALPCLLSLLTQNLKKSIKKEACWTISNITAGNKDQI 362
Query: 797 NFIVTRQALPTLARLIHHSDEEVLTDACWA 886
++ + L L+ ++ ++ +A WA
Sbjct: 363 QAVINAGIIGPLVNLLQTAEFDIKKEAAWA 392
Score = 36.2 bits (82), Expect = 0.75
Identities = 29/115 (25%), Positives = 51/115 (44%), Gaps = 2/115 (1%)
Frame = +2
Query: 413 IEEVIKTGVIPKFVEFLQRHDTPQLQFEAAWALTNVASGTS-EHTKVVIDHNAVPIFVQL 589
I+ VI G+I V LQ + ++ EAAWA++N SG S + K ++ + L
Sbjct: 362 IQAVINAGIIGPLVNLLQTAEF-DIKKEAAWAISNATSGGSHDQIKYLVSEGCIKPLCDL 420
Query: 590 LASPNDDVREQAVWALGNIAGDSPKCRDYVLGH-NALPPLLEQLKENSKISMLRN 751
L P+ + + L NI + G N ++++ + KI L++
Sbjct: 421 LICPDIRIVTVCLEGLENILKVGETDKTLAAGDVNVFSQMIDEAEGLEKIENLQS 475