Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002893A_C01 KCC002893A_c01
(607 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
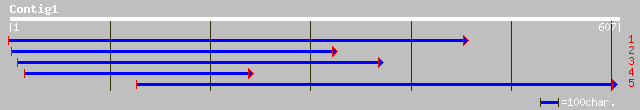
gb|AAM92801.1| hypothetical protein [Oryza sativa (japonica cult... 42 0.009
ref|NP_643836.1| iron receptor [Xanthomonas axonopodis pv. citri... 41 0.012
ref|XP_321223.1| ENSANGP00000020069 [Anopheles gambiae] gi|21289... 37 0.22
pir||I47109 high-sulfur wool matrix protein B2C - sheep gi|94035... 37 0.22
pir||I47112 high-sulfur wool matrix protein B2C - sheep gi|94036... 37 0.22
>gb|AAM92801.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|31430609|gb|AAP52497.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 247
Score = 41.6 bits (96), Expect = 0.009
Identities = 31/93 (33%), Positives = 44/93 (46%), Gaps = 9/93 (9%)
Frame = +1
Query: 148 PLCNLS*TAGSACARQRRRG---------AAQQRCAGPLTGAMLCAQRRCTTTQGPATAG 300
PL +L + + RQ RRG A +RC ++ ++RR T+ P G
Sbjct: 3 PLLHLPPSTRATRQRQVRRGHASSRRLSRAPSRRCPAARPASVAPSRRRPVPTRRPCRVG 62
Query: 301 KPRPRARPSRPYSSSLPQATGRPCVTTLRPPTS 399
+P PRA SRP ++SL A+ RP PP S
Sbjct: 63 RPPPRA-SSRPVAASL--ASSRPAAVDAPPPPS 92
>ref|NP_643836.1| iron receptor [Xanthomonas axonopodis pv. citri str. 306]
gi|21109898|gb|AAM38372.1| iron receptor [Xanthomonas
axonopodis pv. citri str. 306]
Length = 679
Score = 41.2 bits (95), Expect = 0.012
Identities = 48/161 (29%), Positives = 62/161 (37%), Gaps = 2/161 (1%)
Frame = +1
Query: 124 CKVSYAYCPLCNLS*TAGSAC--ARQRRRGAAQQRCAGPLTGAMLCAQRRCTTTQGPATA 297
C+++ A C N + A ++ AR + R +QRC C+ R T PAT
Sbjct: 447 CRLARAMCNWTNAASIATASWNDARAKTRCCRRQRC---------CSSRSRTCRCMPATP 497
Query: 298 GKPRPRARPSRPYSSSLPQATGRPCVTTLRPPTSRMRQH*SWQVDAKSSCLKT*WNR*CS 477
RP AR LP R C RPP MR SW+ +S+C W R CS
Sbjct: 498 RAWRPAAR-----RRGLPITPTRSC----RPP---MRT--SWKPAQRSTCRACAWARHCS 543
Query: 478 ESPRATFWIPTLSGT*SSTHRHPSRRFRG*CWGPTSTCSAC 600
S R T P+RR P + CS C
Sbjct: 544 TSARLT--------------SSPNRR-------PMAVCSTC 563
>ref|XP_321223.1| ENSANGP00000020069 [Anopheles gambiae] gi|21289218|gb|EAA01511.1|
ENSANGP00000020069 [Anopheles gambiae str. PEST]
Length = 260
Score = 37.0 bits (84), Expect = 0.22
Identities = 31/82 (37%), Positives = 34/82 (40%), Gaps = 16/82 (19%)
Frame = +2
Query: 170 RPGRHAHVNGVAALLSSAAPAR*PELCCAHSGAVPQP--------------RAPRQRESQ 307
RP R A + AA+ P P A SG V P RAP R+
Sbjct: 158 RPARAAPKSAAAAVALLLPPVPAPVPSAAGSGRVRLPGAPSPPEGVRGDHGRAPAGRDGS 217
Query: 308 GRGRGRRDHTQARCRR--RPGG 367
GR RGRR RCRR RPGG
Sbjct: 218 GRWRGRR--RSGRCRRFLRPGG 237
>pir||I47109 high-sulfur wool matrix protein B2C - sheep
gi|940359|gb|AAB01448.1| high-sulphur wool matrix
protein B2C
Length = 152
Score = 37.0 bits (84), Expect = 0.22
Identities = 30/110 (27%), Positives = 41/110 (37%), Gaps = 19/110 (17%)
Frame = +1
Query: 124 CKVSYAYCPLCNLS*TAGSACARQRRRGAAQQRCAGPLTGAMLCAQRRCTTTQGPAT--- 294
C S+ P+C+ + T GS+C R +Q C P + C Q C T G T
Sbjct: 4 CSTSFCGFPICSTAGTCGSSCCRST---CSQTSCCQPTSIQTSCCQPTCLQTSGCETGCG 60
Query: 295 ------------AGKPRPRARPSRP----YSSSLPQATGRPCVTTLRPPT 396
+G R R RP +SLP CV + PP+
Sbjct: 61 IGGSTGYGQVGSSGAVSSRTRWCRPDCRVEGTSLPPC----CVVSCTPPS 106
>pir||I47112 high-sulfur wool matrix protein B2C - sheep
gi|940362|gb|AAB01451.1| high-sulphur wool matrix
protein B2C
Length = 152
Score = 37.0 bits (84), Expect = 0.22
Identities = 30/110 (27%), Positives = 41/110 (37%), Gaps = 19/110 (17%)
Frame = +1
Query: 124 CKVSYAYCPLCNLS*TAGSACARQRRRGAAQQRCAGPLTGAMLCAQRRCTTTQGPAT--- 294
C S+ P+C+ + T GS+C R +Q C P + C Q C T G T
Sbjct: 4 CSTSFCGFPICSTAGTCGSSCCRST---CSQTSCCQPTSIQTSCCQPTCLQTSGCETGCG 60
Query: 295 ------------AGKPRPRARPSRP----YSSSLPQATGRPCVTTLRPPT 396
+G R R RP +SLP CV + PP+
Sbjct: 61 IGGSIGYGQVGSSGAVSSRTRWCRPDCRVEGTSLPPC----CVVSCTPPS 106