Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002787A_C01 KCC002787A_c01
(1111 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
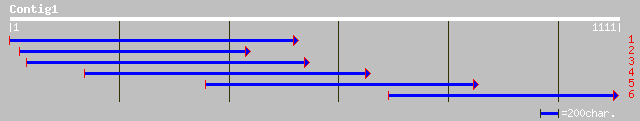
ref|NP_906519.1| conserved hypothetical protein [Wolinella succi... 92 5e-24
ref|NP_207304.1| conserved hypothetical protein [Helicobacter py... 99 5e-22
ref|NP_733202.1| CG31063-PA [Drosophila melanogaster] gi|2318002... 106 8e-22
ref|NP_223175.1| putative [Helicobacter pylori J99] gi|7464728|p... 99 2e-21
gb|AAD15563.1| unknown [Homo sapiens] 72 4e-20
>ref|NP_906519.1| conserved hypothetical protein [Wolinella succinogenes]
gi|34482418|emb|CAE09419.1| conserved hypothetical
protein [Wolinella succinogenes]
Length = 190
Score = 91.7 bits (226), Expect(2) = 5e-24
Identities = 49/126 (38%), Positives = 72/126 (56%), Gaps = 1/126 (0%)
Frame = +2
Query: 296 SSMYITPKAAMYNFRGKERRWDVVESHASVGVVLYHKDLDAWIIVRQFRPAVYATLMREA 475
+S Y+ PK Y RG E+ WDVVE+H SV V+L+ A+++V+QFRPAV+
Sbjct: 12 NSPYVKPKRVFYTERGIEKSWDVVEAHDSVSVLLFDPKKRAFVLVKQFRPAVFL------ 65
Query: 476 EAAGKPKPPYSGAFTYELCAGLIDK-DKSNPEICKEEIMEECGFDVPLEGIHEIGVGIAS 652
+ +TYELCAG++DK KS I +EEI+EECG+ + ++ + I S
Sbjct: 66 --------KGTSGYTYELCAGIVDKPGKSLEVIAQEEILEECGYSLSIDSLERITSFYTS 117
Query: 653 AGTPGA 670
G G+
Sbjct: 118 VGFAGS 123
Score = 42.7 bits (99), Expect(2) = 5e-24
Identities = 24/56 (42%), Positives = 34/56 (59%)
Frame = +1
Query: 691 AEVDSSMAVAGAGGGLLGHGECIEVLALPFES*QQFVLDGKLPKSPGLMFGITWAY 858
AEV+ S+ G GGG+ E IEV+ LP + F+LD + PK+PGL+F W +
Sbjct: 130 AEVEDSLR-QGEGGGV--ESENIEVVHLPLLEARAFMLDEERPKTPGLLFAFMWFF 182
>ref|NP_207304.1| conserved hypothetical protein [Helicobacter pylori 26695]
gi|7463977|pir||C64583 conserved hypothetical protein
HP0507 - Helicobacter pylori (strain 26695)
gi|2313618|gb|AAD07572.1| conserved hypothetical protein
[Helicobacter pylori 26695]
Length = 212
Score = 99.4 bits (246), Expect(2) = 5e-22
Identities = 56/137 (40%), Positives = 81/137 (58%), Gaps = 1/137 (0%)
Frame = +2
Query: 266 ESSFRLEPLESSMYITPKAAMYNFRGKERRWDVVESHASVGVVLYHKDLDAWIIVRQFRP 445
+SS LEP SS +I K YN ++ WD+++S SV V+LY K+ D ++IV+QFRP
Sbjct: 16 DSSVYLEPCSSSNFIELKRMHYNEENTKKTWDIIKSLDSVAVLLYEKESDCFVIVKQFRP 75
Query: 446 AVYATLMREAEAAGKPKPPYSGAFTYELCAGLIDK-DKSNPEICKEEIMEECGFDVPLEG 622
A+YA R G +TYELCAGL+DK +KS EI EE +EECG+ + +
Sbjct: 76 AIYA---RRFHFKCDQDQTIDG-YTYELCAGLVDKANKSLEEIACEEALEECGYQISPKN 131
Query: 623 IHEIGVGIASAGTPGAV 673
+ IG ++ G G++
Sbjct: 132 LETIGQFYSATGLSGSL 148
Score = 28.5 bits (62), Expect(2) = 5e-22
Identities = 20/54 (37%), Positives = 28/54 (51%)
Frame = +1
Query: 691 AEVDSSMAVAGAGGGLLGHGECIEVLALPFES*QQFVLDGKLPKSPGLMFGITW 852
AEV ++ V+ GGG+ E IEVL L F++D + K+ GL I W
Sbjct: 154 AEVHKNLKVS-KGGGI--DTERIEVLFLERSKALDFIMDFQYAKTTGLSLAILW 204
>ref|NP_733202.1| CG31063-PA [Drosophila melanogaster] gi|23180025|gb|AAF56679.2|
CG31063-PA [Drosophila melanogaster]
Length = 404
Score = 106 bits (264), Expect = 8e-22
Identities = 60/140 (42%), Positives = 84/140 (59%), Gaps = 6/140 (4%)
Frame = +2
Query: 269 SSFRLEPL-ESSMYITPKAAMYNFRGKERRWDVVESHASVGVVLYHKDLDAWIIVRQFRP 445
S L PL + S Y+ P Y G E+ WD+++ H SV ++LY+ ++VRQFRP
Sbjct: 197 SKIWLGPLPQDSPYVKPFRLYYVQNGVEKNWDLLKVHDSVAIILYNTSRQKLVLVRQFRP 256
Query: 446 AVYATLMREA-----EAAGKPKPPYSGAFTYELCAGLIDKDKSNPEICKEEIMEECGFDV 610
AVY ++ A E K PP G T ELCAG++DK+KS EI +EE++EECG+DV
Sbjct: 257 AVYHGIISSAKGTFDEVDLKEFPPAIGV-TLELCAGIVDKNKSWVEIAREEVVEECGYDV 315
Query: 611 PLEGIHEIGVGIASAGTPGA 670
P+E I E+ V + G+ GA
Sbjct: 316 PVERIEEVMVYRSGVGSSGA 335
Score = 84.7 bits (208), Expect = 3e-15
Identities = 51/133 (38%), Positives = 75/133 (56%), Gaps = 12/133 (9%)
Frame = +2
Query: 293 ESSMYITPKAAMYNFRGKERRWDVVESHASVGVVLYHKDLDAWIIVRQFRPAVYATL--- 463
+ S +I P Y E++ D++++ V V+LY+K + I VRQFR AVY +
Sbjct: 3 KDSNWIKPGRLHYIENDVEKQVDIIKTIDGVVVILYNKAREKLIFVRQFRGAVYQGIHSA 62
Query: 464 ----MREAEAAGKPKPPYSGAFTYELCAGLIDKDKSNPEICKEEIMEECGFDVPLEGIHE 631
M + EA + PP G T ELC G +DKDKS EI KEE++EECG++VP E +
Sbjct: 63 GSPDMSKGEADLEQFPPEVGV-TLELCGGAVDKDKSLAEIAKEEVLEECGYEVPTESLQH 121
Query: 632 I-----GVGIASA 655
+ G+G +S+
Sbjct: 122 VYDYRSGIGTSSS 134
>ref|NP_223175.1| putative [Helicobacter pylori J99] gi|7464728|pir||F71928
hypothetical protein jhp0457 - Helicobacter pylori
(strain J99) gi|4154997|gb|AAD06038.1| putative
[Helicobacter pylori J99]
Length = 212
Score = 99.0 bits (245), Expect(2) = 2e-21
Identities = 54/137 (39%), Positives = 81/137 (58%), Gaps = 1/137 (0%)
Frame = +2
Query: 266 ESSFRLEPLESSMYITPKAAMYNFRGKERRWDVVESHASVGVVLYHKDLDAWIIVRQFRP 445
+SS LEP SS +I K YN ++ WD+++S SV V+LY K+ D ++IV+QFRP
Sbjct: 16 DSSVYLEPCSSSNFIELKRMHYNEENTKKTWDIIKSLDSVAVLLYEKESDCFVIVKQFRP 75
Query: 446 AVYATLMREAEAAGKPKPPYSGAFTYELCAGLIDK-DKSNPEICKEEIMEECGFDVPLEG 622
A+YA + + +TYELCAGL+DK +KS EI EE +EECG+ + +
Sbjct: 76 AIYARNF----YFKRDQDQTIDGYTYELCAGLVDKANKSLEEIACEEALEECGYQISPKN 131
Query: 623 IHEIGVGIASAGTPGAV 673
+ IG ++ G G++
Sbjct: 132 LETIGQFYSATGLSGSL 148
Score = 26.6 bits (57), Expect(2) = 2e-21
Identities = 19/54 (35%), Positives = 26/54 (47%)
Frame = +1
Query: 691 AEVDSSMAVAGAGGGLLGHGECIEVLALPFES*QQFVLDGKLPKSPGLMFGITW 852
AE + V+ GGG+ E IEVL L F++D + K+ GL I W
Sbjct: 154 AEAHEGLKVS-KGGGI--DTEKIEVLFLERSKALDFIMDFQYAKTTGLSLAILW 204
>gb|AAD15563.1| unknown [Homo sapiens]
Length = 290
Score = 71.6 bits (174), Expect(2) = 4e-20
Identities = 50/191 (26%), Positives = 85/191 (44%), Gaps = 13/191 (6%)
Frame = +2
Query: 137 ACKASSAAAEMAGPIKYPNMYPYGVKTLTPGQTVVEALDMLGPESSFRLEPLESSMYITP 316
AC + GP+ + P + L A++ + S R +S Y+ P
Sbjct: 32 ACTRHARVRAYPGPLVHRRKRPAWLWELAAPACPGAAMERIEGASVGRCA---ASPYLRP 88
Query: 317 KAAMYNFRGKERRWDVVESHASVGVVLYHKDLDAWIIVRQFRPAVY------------AT 460
Y G ++ WD +++H SV V+L++ + ++V+QFRPAVY A
Sbjct: 89 LTLHYRQNGAQKSWDFMKTHDSVTVLLFNSSRRSLVLVKQFRPAVYAGEVERRFPGSLAA 148
Query: 461 LMREAEAAGKPKPPYSGAFTYELCAGLIDK-DKSNPEICKEEIMEECGFDVPLEGIHEIG 637
+ ++ +P P S T ELCAGL+D+ S E+ +E EECG+ + + +
Sbjct: 149 VDQDGPRELQPALPGSAGVTVELCAGLVDQPGLSLEEVACKEAWEECGYHLAPSDLRRVA 208
Query: 638 VGIASAGTPGA 670
+ G G+
Sbjct: 209 TYWSGVGLTGS 219
Score = 49.7 bits (117), Expect(2) = 4e-20
Identities = 22/45 (48%), Positives = 31/45 (68%)
Frame = +1
Query: 718 AGAGGGLLGHGECIEVLALPFES*QQFVLDGKLPKSPGLMFGITW 852
+G GGGL+ GE IEV+ LP E Q F D +PK+ G++FG++W
Sbjct: 234 SGPGGGLVEEGELIEVVHLPLEGAQAFADDPDIPKTLGVIFGVSW 278