Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002749A_C01 KCC002749A_c01
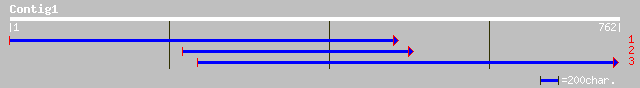
(761 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P01399|TXWB_NAJHA Weak toxin CM-13b gi|69431|pir||V6NJ3E veno... 36 0.58
sp|P25680|TXW0_NAJNI Weak toxin CM-10 35 0.99
gb|AAO27562.1| envelope glycoprotein gpUL73 [human herpesvirus 5] 35 1.3
gb|AAG23514.1| structural glycoprotein gpUL73 [human herpesvirus... 35 1.3
dbj|BAC86651.1| unnamed protein product [Homo sapiens] 35 1.3
>sp|P01399|TXWB_NAJHA Weak toxin CM-13b gi|69431|pir||V6NJ3E venom protein CM-13b - cobra
(Naja haje annulifera) gi|229548|prf||754241A toxin
CM13b
Length = 65
Score = 36.2 bits (82), Expect = 0.58
Identities = 19/60 (31%), Positives = 27/60 (44%), Gaps = 4/60 (6%)
Frame = +2
Query: 278 PMLYSSSERTARNGLTVCW----SESLCGKDVQRRCTVRCIIAAPSRLLHVLECCAAVLC 445
P +Y + T RNG +C+ L GK R C C +A P ++ECC+ C
Sbjct: 7 PEVYCNRFHTCRNGEKICFKRFNERKLLGKRYTRGCAATCPVAKPR---EIVECCSTDRC 63
>sp|P25680|TXW0_NAJNI Weak toxin CM-10
Length = 65
Score = 35.4 bits (80), Expect = 0.99
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 4/60 (6%)
Frame = +2
Query: 278 PMLYSSSERTARNGLTVCW----SESLCGKDVQRRCTVRCIIAAPSRLLHVLECCAAVLC 445
P ++ + T RNG +C+ L GK R C V C +A P ++ECC+ C
Sbjct: 7 PEVFCRNFHTCRNGEKICFKRFDQRKLLGKRYTRGCAVTCPVAKPR---EIVECCSTDGC 63
>gb|AAO27562.1| envelope glycoprotein gpUL73 [human herpesvirus 5]
Length = 134
Score = 35.0 bits (79), Expect = 1.3
Identities = 24/68 (35%), Positives = 32/68 (46%), Gaps = 6/68 (8%)
Frame = +2
Query: 497 VLASVVTESLGTKSVATPHTRQQTHESCG-QGWTIARECALTYTHTQHTHGRRPG---HP 664
++ SVV S T + +TP TH S + T+A T T T T +PG H
Sbjct: 11 LVLSVVASSNNTSTASTPRPSSSTHASTTVKATTVATTSTTTATSTSSTTSAKPGSTTHD 70
Query: 665 PD--RPHA 682
P+ RPHA
Sbjct: 71 PNVMRPHA 78
>gb|AAG23514.1| structural glycoprotein gpUL73 [human herpesvirus 5]
gi|10801794|gb|AAG23515.1| structural glycoprotein
gpUL73 [human herpesvirus 5] gi|10801796|gb|AAG23516.1|
structural glycoprotein gpUL73 [human herpesvirus 5]
gi|10801798|gb|AAG23517.1| structural glycoprotein
gpUL73 [human herpesvirus 5] gi|10801800|gb|AAG23518.1|
structural glycoprotein gpUL73 [human herpesvirus 5]
gi|10801802|gb|AAG23519.1| structural glycoprotein
gpUL73 [human herpesvirus 5] gi|10801804|gb|AAG23520.1|
structural glycoprotein gpUL73 [human herpesvirus 5]
gi|18656479|gb|AAL77787.1| structural glycoprotein UL73
[human herpesvirus 5] gi|18656481|gb|AAL77788.1|
structural glycoprotein UL73 [human herpesvirus 5]
gi|18656483|gb|AAL77789.1| structural glycoprotein UL73
[human herpesvirus 5] gi|18656485|gb|AAL77790.1|
structural glycoprotein UL73 [human herpesvirus 5]
gi|18656487|gb|AAL77791.1| structural glycoprotein UL73
[human herpesvirus 5] gi|18656489|gb|AAL77792.1|
structural glycoprotein UL73 [human herpesvirus 5]
gi|18656491|gb|AAL77793.1| structural glycoprotein UL73
[human herpesvirus 5]
gi|21951878|gb|AAM82385.1|AF390759_1 structural
glycoprotein gpUL73 [human herpesvirus 5]
gi|21951886|gb|AAM82389.1|AF390763_1 structural
glycoprotein gpUL73 [human herpesvirus 5]
gi|21951888|gb|AAM82390.1|AF390764_1 structural
glycoprotein gpUL73 [human herpesvirus 5]
gi|21951902|gb|AAM82397.1|AF390771_1 structural
glycoprotein gpUL73 [human herpesvirus 5]
gi|21951904|gb|AAM82398.1|AF390772_1 structural
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818208|gb|AAO24837.1|AF390798_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818212|gb|AAO24839.1|AF390800_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818218|gb|AAO24842.1|AF390803_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818226|gb|AAO24846.1|AF390807_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818228|gb|AAO24847.1|AF390808_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818230|gb|AAO24848.1|AF390809_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818234|gb|AAO24850.1|AF390811_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818256|gb|AAO24861.1|AF390822_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818260|gb|AAO24863.1|AF390824_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818268|gb|AAO24867.1|AF390828_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818278|gb|AAO24872.1|AF390833_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27818292|gb|AAO24879.1|AF390840_1 envelope
glycoprotein gpUL73 [human herpesvirus 5]
gi|27923291|gb|AAO27549.1| envelope glycoprotein gpUL73
[human herpesvirus 5] gi|27923313|gb|AAO27560.1|
envelope glycoprotein gpUL73 [human herpesvirus 5]
gi|27923321|gb|AAO27564.1| envelope glycoprotein gpUL73
[human herpesvirus 5] gi|28076649|gb|AAO31531.1|
envelope glycoprotein gpUL73 [human herpesvirus 5]
gi|28076657|gb|AAO31535.1| envelope glycoprotein gpUL73
[human herpesvirus 5]
Length = 135
Score = 35.0 bits (79), Expect = 1.3
Identities = 24/68 (35%), Positives = 32/68 (46%), Gaps = 6/68 (8%)
Frame = +2
Query: 497 VLASVVTESLGTKSVATPHTRQQTHESCG-QGWTIARECALTYTHTQHTHGRRPG---HP 664
++ SVV S T + +TP TH S + T+A T T T T +PG H
Sbjct: 11 LVLSVVASSNNTSTASTPRPSSSTHASTTVKATTVATTSTTTATSTSSTTSAKPGSTTHD 70
Query: 665 PD--RPHA 682
P+ RPHA
Sbjct: 71 PNVMRPHA 78
>dbj|BAC86651.1| unnamed protein product [Homo sapiens]
Length = 183
Score = 35.0 bits (79), Expect = 1.3
Identities = 27/71 (38%), Positives = 31/71 (43%), Gaps = 7/71 (9%)
Frame = -3
Query: 663 GWPGLRPCV-----CCVCVYVSAHSLAIVHP-CPQLS*VCCRV-CGVATLLVPKDSVTTL 505
GWPGLR C+ CC V S+ SL + P C S C + CG A L P SV
Sbjct: 42 GWPGLRTCLPDSAPCCRAVTPSSQSLGPLRPLCASCSQPGCSISCGTAAL--PTQSVVPP 99
Query: 504 AKTYSSLAQAL 472
SL L
Sbjct: 100 PSPLLSLPPTL 110