Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002565A_C01 KCC002565A_c01
(654 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
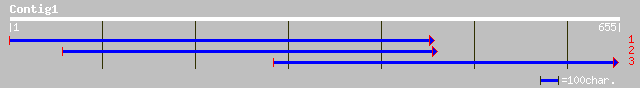
pir||T47988 serine/threonine-protein kinase-like protein - Arabi... 124 3e-46
ref|NP_567122.1| protein kinase family [Arabidopsis thaliana] gi... 124 3e-46
gb|AAO01099.1| CG10967-PA [Drosophila littoralis] 108 4e-37
dbj|BAC65613.1| mKIAA0623 protein [Mus musculus] 107 9e-37
ref|NP_038909.2| Unc-51 like kinase 2 [Mus musculus] gi|4760561|... 107 9e-37
>pir||T47988 serine/threonine-protein kinase-like protein - Arabidopsis thaliana
gi|6899894|emb|CAB71903.1| serine/threonine-protein
kinase-like protein [Arabidopsis thaliana]
Length = 648
Score = 124 bits (311), Expect(2) = 3e-46
Identities = 58/109 (53%), Positives = 75/109 (68%), Gaps = 1/109 (0%)
Frame = +2
Query: 269 SPQCMAETLCGSPLYMAPEVLQLARYDAKADLWSVGTILFELLAGRPPFQGANHLQLVQN 448
+P+ MAET CGSPLYMAPE+++ +YDAKADLWS G ILF+L+ G+PPF G NH+QL N
Sbjct: 161 TPESMAETFCGSPLYMAPEIIRNQKYDAKADLWSAGAILFQLVTGKPPFDGNNHIQLFHN 220
Query: 449 IERG-DAVLPDAVARALTPGCRQLLYQLLRRNPVERISHDELFAHPFLQ 592
I R + P+ + P C L LLRRNP+ER++ E F H FL+
Sbjct: 221 IVRDTELKFPEDTRNEIHPDCVDLCRSLLRRNPIERLTFREFFNHMFLR 269
Score = 83.2 bits (204), Expect(2) = 3e-46
Identities = 47/84 (55%), Positives = 52/84 (60%)
Frame = +1
Query: 22 KIFLVLEYCGGGDLAQYLRHRGPAERGCRAWYLLRHLA*WAQCAAVAHNIIHRDLKPQNL 201
+IFLVLEYC GGDLA Y+ G A + +R LA Q H IHRDLKPQNL
Sbjct: 81 RIFLVLEYCSGGDLAGYINRHGKVPEAV-AKHFMRQLALGLQVLQEKH-FIHRDLKPQNL 138
Query: 202 LLSDSGPSPTLKIADFGFARSLQP 273
LLS +P LKI DFGFARSL P
Sbjct: 139 LLSSKEVTPLLKIGDFGFARSLTP 162
>ref|NP_567122.1| protein kinase family [Arabidopsis thaliana]
gi|14334752|gb|AAK59554.1| putative
serine/threonine-protein kinase [Arabidopsis thaliana]
Length = 626
Score = 124 bits (311), Expect(2) = 3e-46
Identities = 58/109 (53%), Positives = 75/109 (68%), Gaps = 1/109 (0%)
Frame = +2
Query: 269 SPQCMAETLCGSPLYMAPEVLQLARYDAKADLWSVGTILFELLAGRPPFQGANHLQLVQN 448
+P+ MAET CGSPLYMAPE+++ +YDAKADLWS G ILF+L+ G+PPF G NH+QL N
Sbjct: 161 TPESMAETFCGSPLYMAPEIIRNQKYDAKADLWSAGAILFQLVTGKPPFDGNNHIQLFHN 220
Query: 449 IERG-DAVLPDAVARALTPGCRQLLYQLLRRNPVERISHDELFAHPFLQ 592
I R + P+ + P C L LLRRNP+ER++ E F H FL+
Sbjct: 221 IVRDTELKFPEDTRNEIHPDCVDLCRSLLRRNPIERLTFREFFNHMFLR 269
Score = 83.2 bits (204), Expect(2) = 3e-46
Identities = 47/84 (55%), Positives = 52/84 (60%)
Frame = +1
Query: 22 KIFLVLEYCGGGDLAQYLRHRGPAERGCRAWYLLRHLA*WAQCAAVAHNIIHRDLKPQNL 201
+IFLVLEYC GGDLA Y+ G A + +R LA Q H IHRDLKPQNL
Sbjct: 81 RIFLVLEYCSGGDLAGYINRHGKVPEAV-AKHFMRQLALGLQVLQEKH-FIHRDLKPQNL 138
Query: 202 LLSDSGPSPTLKIADFGFARSLQP 273
LLS +P LKI DFGFARSL P
Sbjct: 139 LLSSKEVTPLLKIGDFGFARSLTP 162
>gb|AAO01099.1| CG10967-PA [Drosophila littoralis]
Length = 844
Score = 108 bits (269), Expect(2) = 4e-37
Identities = 57/116 (49%), Positives = 77/116 (66%), Gaps = 1/116 (0%)
Frame = +2
Query: 281 MAETLCGSPLYMAPEVLQLARYDAKADLWSVGTILFELLAGRPPFQGANHLQLVQNIERG 460
MA TLCGSP+YMAPEV+ +YDAKADLWS+GTI+++ L G+ PF +L E+
Sbjct: 173 MAATLCGSPMYMAPEVIMSLQYDAKADLWSLGTIVYQCLTGKAPFYAQTPHELKFYYEQ- 231
Query: 461 DAVLPDAVARALTPGCRQLLYQLLRRNPVERISHDELFAHPFLQG-EAASAAVQLP 625
+A L + ++P R LL LLRRN +RIS++ F HPFLQG +AA + V +P
Sbjct: 232 NANLAPKIPHGVSPDLRDLLLCLLRRNSKDRISYESFFVHPFLQGKKAAVSPVDMP 287
Score = 68.9 bits (167), Expect(2) = 4e-37
Identities = 43/90 (47%), Positives = 52/90 (57%), Gaps = 7/90 (7%)
Frame = +1
Query: 31 LVLEYCGGGDLAQYLRHRGPAERGCRAWYLLRHLA*WAQCAAVAHNIIHRDLKPQNLLLS 210
LV+EYC GGDLA YL +G +L++ A A I+HRDLKPQN+LLS
Sbjct: 85 LVMEYCNGGDLADYLSVKGTLSEDTVRLFLIQLAG--AMKALYTKGIVHRDLKPQNILLS 142
Query: 211 DSG----PSP---TLKIADFGFARSLQPAV 279
+ P+P TLKIADFGFAR L V
Sbjct: 143 HNYGKTLPAPSKITLKIADFGFARFLNEGV 172
>dbj|BAC65613.1| mKIAA0623 protein [Mus musculus]
Length = 1056
Score = 107 bits (267), Expect(2) = 9e-37
Identities = 50/118 (42%), Positives = 75/118 (63%)
Frame = +2
Query: 281 MAETLCGSPLYMAPEVLQLARYDAKADLWSVGTILFELLAGRPPFQGANHLQLVQNIERG 460
MA TLCGSP+YMAPEV+ YDAKADLWS+GT++++ L G+PPFQ + L E+
Sbjct: 189 MAATLCGSPMYMAPEVIMSQHYDAKADLWSIGTVIYQCLVGKPPFQANSPQDLRMFYEKN 248
Query: 461 DAVLPDAVARALTPGCRQLLYQLLRRNPVERISHDELFAHPFLQGEAASAAVQLPAPI 634
+++P ++ R +P LL LL+RN +R+ + F+HPFL+ + +P P+
Sbjct: 249 RSLMP-SIPRETSPYLANLLLGLLQRNQKDRMDFEAFFSHPFLEQVPVKKSCPVPVPV 305
Score = 68.6 bits (166), Expect(2) = 9e-37
Identities = 42/95 (44%), Positives = 54/95 (56%), Gaps = 6/95 (6%)
Frame = +1
Query: 1 DLFKEPGKIFLVLEYCGGGDLAQYLRHRGPAERGCRAWYLLRHLA*WAQCAAVAHNIIHR 180
D+ + P +FLV+EYC GGDLA YL+ +G +L H A + IIHR
Sbjct: 92 DVQELPNSVFLVMEYCNGGDLADYLQAKGTLSEDTIRVFL--HQIAAAMRILHSKGIIHR 149
Query: 181 DLKPQNLLLSDSGPSPT------LKIADFGFARSL 267
DLKPQN+LLS + + +KIADFGFAR L
Sbjct: 150 DLKPQNILLSYANRRKSNVSGIRIKIADFGFARYL 184
>ref|NP_038909.2| Unc-51 like kinase 2 [Mus musculus] gi|4760561|dbj|BAA77341.1|
UNC-51-like kinase (ULK) 2 [Mus musculus]
Length = 1037
Score = 107 bits (267), Expect(2) = 9e-37
Identities = 50/118 (42%), Positives = 75/118 (63%)
Frame = +2
Query: 281 MAETLCGSPLYMAPEVLQLARYDAKADLWSVGTILFELLAGRPPFQGANHLQLVQNIERG 460
MA TLCGSP+YMAPEV+ YDAKADLWS+GT++++ L G+PPFQ + L E+
Sbjct: 170 MAATLCGSPMYMAPEVIMSQHYDAKADLWSIGTVIYQCLVGKPPFQANSPQDLRMFYEKN 229
Query: 461 DAVLPDAVARALTPGCRQLLYQLLRRNPVERISHDELFAHPFLQGEAASAAVQLPAPI 634
+++P ++ R +P LL LL+RN +R+ + F+HPFL+ + +P P+
Sbjct: 230 RSLMP-SIPRETSPYLANLLLGLLQRNQKDRMDFEAFFSHPFLEQVPVKKSCPVPVPV 286
Score = 68.6 bits (166), Expect(2) = 9e-37
Identities = 42/95 (44%), Positives = 54/95 (56%), Gaps = 6/95 (6%)
Frame = +1
Query: 1 DLFKEPGKIFLVLEYCGGGDLAQYLRHRGPAERGCRAWYLLRHLA*WAQCAAVAHNIIHR 180
D+ + P +FLV+EYC GGDLA YL+ +G +L H A + IIHR
Sbjct: 73 DVQELPNSVFLVMEYCNGGDLADYLQAKGTLSEDTIRVFL--HQIAAAMRILHSKGIIHR 130
Query: 181 DLKPQNLLLSDSGPSPT------LKIADFGFARSL 267
DLKPQN+LLS + + +KIADFGFAR L
Sbjct: 131 DLKPQNILLSYANRRKSNVSGIRIKIADFGFARYL 165