Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002472A_C01 KCC002472A_c01
(1015 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
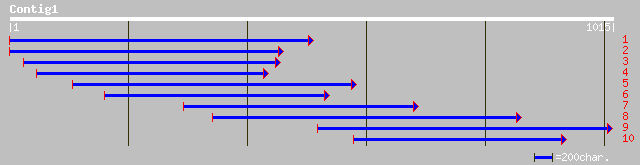
ref|ZP_00056820.1| COG2115: Xylose isomerase [Thermobifida fusca] 53 9e-06
ref|NP_823720.1| hypothetical protein [Streptomyces avermitilis ... 49 1e-04
ref|XP_304245.1| hypothetical protein XP_304245 [Homo sapiens] 48 2e-04
ref|NP_652552.1| CG13990-PA [Drosophila melanogaster] gi|2294574... 48 3e-04
pir||G84693 probable proline-rich protein [imported] - Arabidops... 48 3e-04
>ref|ZP_00056820.1| COG2115: Xylose isomerase [Thermobifida fusca]
Length = 754
Score = 52.8 bits (125), Expect = 9e-06
Identities = 45/175 (25%), Positives = 62/175 (34%), Gaps = 6/175 (3%)
Frame = -1
Query: 706 GAAPQRQRPTPTRACRRSSPEPQSWPRPDPAPGKLQNRRRLS**SWRCRRRSSAPRGCAR 527
GA P Q P+P S+ P + P P P + SS P
Sbjct: 6 GAKPSAQSPSPASPSPNSAGSPNTNPTTPPEP-----------------KASSCP----- 43
Query: 526 T*GPPSRRTGPKTPSP--GRIPRKRARWPLEVPRTPRCNPAKT*AGC*GRKSCGIFPPAA 353
T G P+ P PSP P P P P C+P+ + A R+S +A
Sbjct: 44 TTGSPNALPKPPNPSPTAATPPAPATTTPPPTPTGPTCSPSPSAANSAPRESPPPPKSSA 103
Query: 352 CAERSSSTAPSCGSPKTKARWPCHAGCRGGPA----PCVCADPPGPAPSRRAPQP 200
+ +S + +P WP H+ PA P +PP P+P P P
Sbjct: 104 ASPPPNSPPSTSSAPAPATTWPPHSPSTPDPATSSSPSAPPEPPSPSPKNPPPTP 158
>ref|NP_823720.1| hypothetical protein [Streptomyces avermitilis MA-4680]
gi|29606192|dbj|BAC70255.1| hypothetical protein
[Streptomyces avermitilis MA-4680]
Length = 810
Score = 48.9 bits (115), Expect = 1e-04
Identities = 54/169 (31%), Positives = 65/169 (37%), Gaps = 2/169 (1%)
Frame = -1
Query: 700 APQRQRPTPTRACRRSSPEPQSWPRPDPAPGKLQNRRRLS**SWRCRRRSSAPRGCART* 521
AP+ Q P P++A R S P PR P + +R R S R R R P
Sbjct: 62 APRPQPPLPSKASRSRSRSPSRRPRRASRPRRPGSRSRSPRSSRRHRSRHPRP------- 114
Query: 520 GPPSRRTGPKTPSPGRIPRKRARWPLEVPRTPRCNPAKT*AGC*GRKSCGIFPPAACAER 341
G RR P+ P P R P RAR P P+ G G+ P A
Sbjct: 115 GTSLRRPHPRIPCPRRRPTPRAR-VTGAPAAPQAGY--------GFPQPGV--PTPPAPD 163
Query: 340 SSSTAPSCGSPKTKARWPCHAGCRGGP-APCVCADP-PGPAPSRRAPQP 200
S G P+ A+ AG GP +P A P P APS PQP
Sbjct: 164 SRVKQDGYGFPQAGAQGVPQAGPPAGPISPAAQAAPAPPAAPSAPTPQP 212
>ref|XP_304245.1| hypothetical protein XP_304245 [Homo sapiens]
Length = 208
Score = 48.1 bits (113), Expect = 2e-04
Identities = 53/174 (30%), Positives = 61/174 (34%), Gaps = 35/174 (20%)
Frame = -1
Query: 565 CRRRSSA---PRGCART*G----PPSRRTGPKTPSPGRIPRKRARWPLEVPRTPRC---- 419
CRR + PR CAR P + P TP PG WP PRTP C
Sbjct: 17 CRRSTCCCPRPRPCARVASTCCRPRAAPRCPPTPPPG------PAWP---PRTPTCCWGP 67
Query: 418 -NPAKT*AGC*GRKSCGIFPPAACAERSSSTAPSCGSP-------------KTKARWPCH 281
P AGC R +C C R S P C +P +ARW
Sbjct: 68 GGPQHRCAGCRHRPACTPTTTTPCTARRSR--PGCPTPAGAGAHWPAGRPSSARARWSSC 125
Query: 280 AGCRGGPAPCVCADPPGPAP------SRRAPQPL--CGPQGSVL--GSVQQRRC 149
+ R P CA P S P+PL P G S+QQRRC
Sbjct: 126 SSSRATTCPSTCARPARTRDNLTWMMSSLTPRPLLASSPWGRYFLPVSLQQRRC 179
>ref|NP_652552.1| CG13990-PA [Drosophila melanogaster] gi|22945747|gb|AAF52351.2|
CG13990-PA [Drosophila melanogaster]
Length = 471
Score = 47.8 bits (112), Expect = 3e-04
Identities = 52/231 (22%), Positives = 87/231 (37%)
Frame = +2
Query: 119 AQQEACCTPLAAALLYTT*HRTLGAAKRLRCSPTWSRPRRICANARRWTTSTTCVARPSS 298
A++ C P ++ TT C+PT + CA TT+TTC ++
Sbjct: 114 AEETTTCAP---QIITTTTCTPASVTTTTTCAPTTTTT---CAP----TTTTTCAPTTTT 163
Query: 299 LCFGTSAARCS*T*ALSTSRWWKDATTFSTSTTCSRLCGVAARRTRNF*RPASPFARNTT 478
C T+ C+ T + + +T+TTC+ T +P T
Sbjct: 164 TCAPTTTTTCAPTTTTTCAPTTTTTCAPTTTTTCAPTTTTTCAPTTT--TTCAPTTTTTC 221
Query: 479 W*RSFGSSAPTWRTLSTCATARCTAPSPTAPTLSAQSATILKLSRSWIRPGP*LRLWRTT 658
+ + APT T +TCA T +PT T A + T + P TT
Sbjct: 222 APTTTTTCAPT--TTTTCAPTTTTTCAPTTTTTCAPTTTTTCAPTTTTTCAP---TTTTT 276
Query: 659 AASTRRRWTLPLRRSAWLQPRSPPSSSSAGSRRTSTCCSPTKQSICGASSS 811
A T P + P++++ + T+T C+PT + C +++
Sbjct: 277 CAPTTTTTCAPTTTTT-----CAPTTTTTCAPTTTTTCAPTTTTTCAPTTT 322
Score = 47.4 bits (111), Expect = 4e-04
Identities = 54/220 (24%), Positives = 84/220 (37%)
Frame = +2
Query: 137 CTPLAAALLYTT*HRTLGAAKRLRCSPTWSRPRRICANARRWTTSTTCVARPSSLCFGTS 316
CTP A++ TT T C+PT + CA TT+TTC ++ C T+
Sbjct: 130 CTP--ASVTTTT---TCAPTTTTTCAPTTTTT---CAP----TTTTTCAPTTTTTCAPTT 177
Query: 317 AARCS*T*ALSTSRWWKDATTFSTSTTCSRLCGVAARRTRNF*RPASPFARNTTW*RSFG 496
C+ T + + +T+TTC+ T +P T +
Sbjct: 178 TTTCAPTTTTTCAPTTTTTCAPTTTTTCAPTTTTTCAPTTT--TTCAPTTTTTCAPTTTT 235
Query: 497 SSAPTWRTLSTCATARCTAPSPTAPTLSAQSATILKLSRSWIRPGP*LRLWRTTAASTRR 676
+ APT T +TCA T +PT T A + T + P TT A T
Sbjct: 236 TCAPT--TTTTCAPTTTTTCAPTTTTTCAPTTTTTCAPTTTTTCAP---TTTTTCAPTTT 290
Query: 677 RWTLPLRRSAWLQPRSPPSSSSAGSRRTSTCCSPTKQSIC 796
P + P++++ + T+T C+PT + C
Sbjct: 291 TTCAPTTTTT-----CAPTTTTTCAPTTTTTCAPTTTTTC 325
Score = 46.2 bits (108), Expect = 9e-04
Identities = 55/248 (22%), Positives = 86/248 (34%)
Frame = +2
Query: 137 CTPLAAALLYTT*HRTLGAAKRLRCSPTWSRPRRICANARRWTTSTTCVARPSSLCFGTS 316
C P T T C+PT + CA TT+TTC ++ C T+
Sbjct: 157 CAPTTTTTCAPTTTTTCAPTTTTTCAPTTTTT---CAP----TTTTTCAPTTTTTCAPTT 209
Query: 317 AARCS*T*ALSTSRWWKDATTFSTSTTCSRLCGVAARRTRNF*RPASPFARNTTW*RSFG 496
C+ T + + +T+TTC+ T +P T +
Sbjct: 210 TTTCAPTTTTTCAPTTTTTCAPTTTTTCAPTTTTTCAPTTT--TTCAPTTTTTCAPTTTT 267
Query: 497 SSAPTWRTLSTCATARCTAPSPTAPTLSAQSATILKLSRSWIRPGP*LRLWRTTAASTRR 676
+ APT T +TCA T +PT T A + T + P TT +
Sbjct: 268 TCAPT--TTTTCAPTTTTTCAPTTTTTCAPTTTTTCAPTTTTTCAP------TTTTTCAP 319
Query: 677 RWTLPLRRSAWLQPRSPPSSSSAGSRRTSTCCSPTKQSICGASSSHMQSIRNAYAEHKHT 856
T +P ++++ T++C S T + CG S S + + K
Sbjct: 320 TTTTTCTPGITTTTCTPATTTTCVPETTTSCASSTTTTECGPVSGVSGSSKARPSSVKVR 379
Query: 857 QSRSTRRA 880
+R R A
Sbjct: 380 PARPVRPA 387
>pir||G84693 probable proline-rich protein [imported] - Arabidopsis thaliana
gi|3980411|gb|AAC95214.1| putative proline-rich protein
[Arabidopsis thaliana]
Length = 891
Score = 47.8 bits (112), Expect = 3e-04
Identities = 54/183 (29%), Positives = 77/183 (41%), Gaps = 16/183 (8%)
Frame = -1
Query: 700 APQRQRPTPTR-ACRRSSPEPQSWPRPDPAPGKLQN-------RRRLS**SWRCRRRSSA 545
AP R+R +P+ A RR SP P + R P+P ++ R+R S R RS
Sbjct: 318 APSRRRRSPSPPARRRRSPSPPARRRRSPSPPARRHRSPTPPARQRRSPSPPARRHRSPP 377
Query: 544 PRGCART*GPPSRRTGPKTPSPGRIPRKRARWPLEVPRTPRC-------NPAKT*AGC*G 386
P R+ PP+RR ++PSP P +R R P + R R N +++ G
Sbjct: 378 PARRRRSPSPPARRR--RSPSP---PARRRRSPSPLYRRNRSPSPLYRRNRSRSPLAKRG 432
Query: 385 RKSCGIFPPAACAERSSSTAPSCGSPKTKARWPCHAGCRGGPAPCV-CADPPGPAPSRRA 209
R P+ A T SP + R P + P+P A P P P++R
Sbjct: 433 RSDSPGRSPSPVARLRDPTGARLPSPSIEQRLPSPPVAQRLPSPPPRRAGLPSPPPAQRL 492
Query: 208 PQP 200
P P
Sbjct: 493 PSP 495
Score = 32.7 bits (73), Expect(2) = 2.3
Identities = 30/96 (31%), Positives = 36/96 (37%), Gaps = 11/96 (11%)
Frame = -1
Query: 313 SPKTKARWPCHAGCRGGPAPC---VCADPPG--------PAPSRRAPQPLCGPQGSVLGS 167
SP + R P H G R PAP PP PA RR+P P S
Sbjct: 300 SPIRRHRRPTHEGRRQSPAPSRRRRSPSPPARRRRSPSPPARRRRSPSPPARRHRSPTPP 359
Query: 166 VQQRRCQWCAACLLLSFRGRRQHRFPSTGLRRQGPA 59
+QRR A R+HR P RR+ P+
Sbjct: 360 ARQRRSPSPPA---------RRHRSPPPARRRRSPS 386
Score = 20.8 bits (42), Expect(2) = 2.3
Identities = 12/41 (29%), Positives = 21/41 (50%)
Frame = -3
Query: 500 WTQNSVTRSYSSQKGSLAFRSSSYAALQPRKDVSRLLRSKK 378
W++N ++ S + SS A++ PRK RL S++
Sbjct: 234 WSRNLFVAKIIIERKSRSTSQSSDASISPRK--RRLSNSRR 272