Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002463A_C01 KCC002463A_c01
(578 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
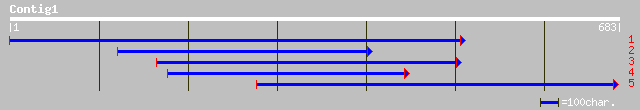
dbj|BAB91333.1| chloroplast ppGpp synthase/degradase [Chlamydomo... 389 e-107
dbj|BAC56909.1| RelA homolog [Suaeda japonica] 82 4e-15
gb|AAL03950.1| relA/spoT-like protein RSH2 [Nicotiana tabacum] 78 1e-13
gb|AAQ23899.1| RSH2 [Nicotiana tabacum] 78 1e-13
gb|AAK82651.1| RSH-like protein [Capsicum annuum] 77 2e-13
>dbj|BAB91333.1| chloroplast ppGpp synthase/degradase [Chlamydomonas reinhardtii]
Length = 735
Score = 389 bits (1000), Expect = e-107
Identities = 191/192 (99%), Positives = 191/192 (99%)
Frame = +1
Query: 1 GYQSLHETVYGEGDVPVEVQIRTHKMHYIAEYGFAAHWKYKEQMDSEDEWLEKETQYKRW 180
GYQSLHETVYGEGDVPVEVQIRTHKMHYIAEYGFAAHWKYKEQMDSEDEWLEKETQYKRW
Sbjct: 483 GYQSLHETVYGEGDVPVEVQIRTHKMHYIAEYGFAAHWKYKEQMDSEDEWLEKETQYKRW 542
Query: 181 LTQYKLRVHDKKVRPQGSPPTDSSLKSLGVAYMDMPEEQQRGLLDPFLQHERFKLQVPAK 360
LTQYKLRVHDKKVRPQGSPPTDSSLKSLGVAYMDMPEEQQRGLLDPFLQHERFKLQVPAK
Sbjct: 543 LTQYKLRVHDKKVRPQGSPPTDSSLKSLGVAYMDMPEEQQRGLLDPFLQHERFKLQVPAK 602
Query: 361 TEVSVLLQTCDGIETKDFPLGTTANQLRRDLGLGAQPGYALTVNSRLPSGEAALQSGDLV 540
TEVSVLLQTCDGIETKDFPLGTTANQL RDLGLGAQPGYALTVNSRLPSGEAALQSGDLV
Sbjct: 603 TEVSVLLQTCDGIETKDFPLGTTANQLWRDLGLGAQPGYALTVNSRLPSGEAALQSGDLV 662
Query: 541 QVLPLSTILSRS 576
QVLPLSTILSRS
Sbjct: 663 QVLPLSTILSRS 674
>dbj|BAC56909.1| RelA homolog [Suaeda japonica]
Length = 708
Score = 82.4 bits (202), Expect = 4e-15
Identities = 53/160 (33%), Positives = 72/160 (44%), Gaps = 5/160 (3%)
Frame = +1
Query: 1 GYQSLHETVYGEGDVPVEVQIRTHKMHYIAEYGFAAHWKYKEQMDSEDEWLEKETQYKRW 180
GYQSLH V GEG VP+EVQIRT +MH AEYGFAAHW+YKE ++ + ++ RW
Sbjct: 487 GYQSLHTVVVGEGMVPLEVQIRTKEMHLQAEYGFAAHWRYKEGESKHSSFVLQMVEWARW 546
Query: 181 LTQYKLRVHDKKVRPQG-----SPPTDSSLKSLGVAYMDMPEEQQRGLLDPFLQHERFKL 345
+ ++ +K G P S G Y P+ G
Sbjct: 547 VVNWQCETMNKDRSSFGYVDSLKSPCKFPTHSEGCPYSYNPQPNHDG------------- 593
Query: 346 QVPAKTEVSVLLQTCDGIETKDFPLGTTANQLRRDLGLGA 465
V V+L + + K+FPL +T L G G+
Sbjct: 594 ------PVFVILIENEKMSVKEFPLNSTMMDLLEATGHGS 627
>gb|AAL03950.1| relA/spoT-like protein RSH2 [Nicotiana tabacum]
Length = 643
Score = 77.8 bits (190), Expect = 1e-13
Identities = 34/65 (52%), Positives = 45/65 (68%)
Frame = +1
Query: 1 GYQSLHETVYGEGDVPVEVQIRTHKMHYIAEYGFAAHWKYKEQMDSEDEWLEKETQYKRW 180
GYQSLH V GEG VP+EVQIRT +MH AEYGFAAHW+YKE ++ + ++ RW
Sbjct: 502 GYQSLHTVVLGEGMVPLEVQIRTKEMHLQAEYGFAAHWRYKEGACKHSSFVNQMVEWARW 561
Query: 181 LTQYK 195
+ ++
Sbjct: 562 VVTWQ 566
>gb|AAQ23899.1| RSH2 [Nicotiana tabacum]
Length = 718
Score = 77.8 bits (190), Expect = 1e-13
Identities = 34/65 (52%), Positives = 45/65 (68%)
Frame = +1
Query: 1 GYQSLHETVYGEGDVPVEVQIRTHKMHYIAEYGFAAHWKYKEQMDSEDEWLEKETQYKRW 180
GYQSLH V GEG VP+EVQIRT +MH AEYGFAAHW+YKE ++ + ++ RW
Sbjct: 502 GYQSLHTVVLGEGMVPLEVQIRTKEMHLQAEYGFAAHWRYKEGACKHSSFVNQMVEWARW 561
Query: 181 LTQYK 195
+ ++
Sbjct: 562 VVTWQ 566
>gb|AAK82651.1| RSH-like protein [Capsicum annuum]
Length = 721
Score = 76.6 bits (187), Expect = 2e-13
Identities = 34/65 (52%), Positives = 45/65 (68%)
Frame = +1
Query: 1 GYQSLHETVYGEGDVPVEVQIRTHKMHYIAEYGFAAHWKYKEQMDSEDEWLEKETQYKRW 180
GYQSLH V GEG VP+EVQIRT +MH AEYGFAAHW+YKE ++ + ++ RW
Sbjct: 502 GYQSLHTVVLGEGMVPLEVQIRTKEMHLQAEYGFAAHWRYKEDDCKHSSFVLQMVEWARW 561
Query: 181 LTQYK 195
+ ++
Sbjct: 562 VVTWQ 566