Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
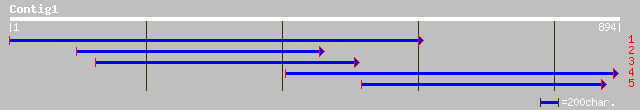
Query= KCC002396A_C01 KCC002396A_c01
(793 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_330656.1| hypothetical protein [Neurospora crassa] gi|289... 53 6e-06
ref|NP_630271.1| proline rich protein (putative membrane protein... 51 2e-05
dbj|BAB85091.1| unnamed protein product [Homo sapiens] 49 9e-05
gb|AAP74957.1| keratinocytes proline-rich protein [Rattus norveg... 48 2e-04
ref|XP_307770.1| ENSANGP00000004103 [Anopheles gambiae] gi|21291... 48 2e-04
>ref|XP_330656.1| hypothetical protein [Neurospora crassa] gi|28927127|gb|EAA36084.1|
hypothetical protein [Neurospora crassa]
Length = 1627
Score = 52.8 bits (125), Expect = 6e-06
Identities = 32/110 (29%), Positives = 51/110 (46%), Gaps = 8/110 (7%)
Frame = +2
Query: 485 PAGATNTEEAFA-VAGSAVGE-------RLAVWRDWQFWRFKLNTTCGADPKQLKYKVDL 640
P G+ N + A + G VG+ RL R + FWRF + +++ Y+++
Sbjct: 723 PPGSFNARKKRARIDGEKVGKYKDVRGFRLHAERGYTFWRFNIEVELREKQQRIAYRINR 782
Query: 641 VPGRAFTVSVPALSEAWHAAFYSCNGLHNPQDYGRTHGIQPLWVDLMRQH 790
P F V PA +A + F+SCNG D + G P+W D++ H
Sbjct: 783 GPATGFWV--PAKGQAMNIMFHSCNGFSLSVDPNQFSGPDPMWRDVLNTH 830
>ref|NP_630271.1| proline rich protein (putative membrane protein) [Streptomyces
coelicolor A3(2)] gi|7479100|pir||T35474 50kD proline
rich protein - Streptomyces coelicolor
gi|4008532|emb|CAA22501.1| proline rich protein
(putative membrane protein) [Streptomyces coelicolor
A3(2)]
Length = 456
Score = 50.8 bits (120), Expect = 2e-05
Identities = 58/183 (31%), Positives = 69/183 (37%), Gaps = 17/183 (9%)
Frame = -3
Query: 791 NAGA*GRPTAAGCRV--CGRSPVGCGGRCRSRR-----RRARPHSAPALTR*RHGRALGP 633
+A A GRP + G R G P G R R R R P P R RH + G
Sbjct: 46 SAAAPGRPWSGGARARHFGARPPTPGDRRRQYRPPHGNRLPEPVPGP---RHRHQQRRGT 102
Query: 632 PCT*----AALDPRRRWY*A*SARTASRATPQAARPLR----CLPRQ-TPPPCWWLQPAQ 480
P A RR A + R A +P+ P R C RQ +P P + +
Sbjct: 103 PAVGTGPEAPASTTRRAGPAPAVRRAGHRSPRPTGPDRKHRTCFSRQPSPSPVSVARASS 162
Query: 479 RSRPQP-PPCTCASKPRPRHCRAPRPTPAAAARCRLGWRTELRCCRPPSPSSGTRRNPHR 303
SRP+P PP +PRP H P P P RPP P R P R
Sbjct: 163 PSRPRPCPPPVPVPRPRPVHDPPPAPAPFLV--------------RPPPPPRPYGRRPPR 208
Query: 302 RRP 294
RRP
Sbjct: 209 RRP 211
>dbj|BAB85091.1| unnamed protein product [Homo sapiens]
Length = 178
Score = 48.9 bits (115), Expect = 9e-05
Identities = 48/155 (30%), Positives = 65/155 (40%), Gaps = 9/155 (5%)
Frame = +3
Query: 336 RWWSAAAQLSPPS*AAACRRSWCGARCPAVARARLGSTRTRRGLRATTLCRLEPPTRRRR 515
RW +A P S A R+ G R PA R ST G + C
Sbjct: 13 RWPTAR----PTSSTAISARTSAG-RTPAGPRRSWRSTSPLPGTASAPCC---------- 57
Query: 516 LPWQAAQW-----ASGLRC----GATGSSGASSSIPPAARIQSSSSTRWT*CPAVPSPCQ 668
PW+A +W A+ C GA G + +SS P + R SS+TRWT CP + +
Sbjct: 58 -PWRAPRWRHSTAATATTCSSSPGARGRARPTSSAPSSPR---SSTTRWTSCPGLSARVT 113
Query: 669 CRR*VRPGTPPSTPATASTTHRTTAAHTASSRCGS 773
RPG P+T +A RT ++S R G+
Sbjct: 114 GAGITRPGL-PTTDRSAQRAPRTRTTSSSSKRPGN 147
>gb|AAP74957.1| keratinocytes proline-rich protein [Rattus norvegicus]
Length = 699
Score = 48.1 bits (113), Expect = 2e-04
Identities = 55/177 (31%), Positives = 63/177 (35%), Gaps = 7/177 (3%)
Frame = -3
Query: 770 PTAAGCRVCGRSPVGCGGRCRSRRRRARPHSAPALTR*RHGRALGPPCT*AALD--PRRR 597
P R C R P G C R A PH+ P PC LD P
Sbjct: 391 PIRGRSRSCPRQP-SWGVSCPDLRPCAEPHAFPR------------PCRPQRLDRSPESS 437
Query: 596 WY*A*SARTASRATPQAARPLRCLPRQTPPPCWWLQPAQRSRPQPPPCTCASKPRPRHCR 417
W R A RP PR P P +P R RP+P PC + +PRPR
Sbjct: 438 W---------RRCPVPAPRPY---PRPEPCPSPEPRPCPRPRPRPEPCP-SPEPRPR--- 481
Query: 416 APRPTPAAAARCRLGWRTELRCCRPPSPSSGTRRNP-----HRRRPV*ERGQTPADR 261
PRP P + R R E C P P R +P R RP E +P R
Sbjct: 482 -PRPDPCPSPELRPRPRPE--PCPSPEPRPRPRPDPCPSPEPRPRPCPEPCPSPEPR 535
Score = 45.4 bits (106), Expect = 0.001
Identities = 37/113 (32%), Positives = 45/113 (39%), Gaps = 7/113 (6%)
Frame = -3
Query: 713 CRSRRRRARPHSAPALTR*RHGRALGPPCT*AALDPRRRWY*A*SARTASRATPQ---AA 543
C S R RP P + R PC PR R S R P+ +
Sbjct: 473 CPSPEPRPRPRPDPCPSPELRPRPRPEPCPSPEPRPRPRPDPCPSPEPRPRPCPEPCPSP 532
Query: 542 RPLRCLP-RQTPPPCWWLQPAQRSRPQPPPCTC-ASKPRPRHCRAP--RPTPA 396
P C P R+ PC + +P S+P P P C A PRP HC P RP P+
Sbjct: 533 EPRPCPPLRRFSEPCLYPEPCSVSKPVPCPVPCPAPHPRPVHCETPGRRPQPS 585
>ref|XP_307770.1| ENSANGP00000004103 [Anopheles gambiae] gi|21291398|gb|EAA03543.1|
ENSANGP00000004103 [Anopheles gambiae str. PEST]
Length = 972
Score = 48.1 bits (113), Expect = 2e-04
Identities = 39/127 (30%), Positives = 48/127 (37%), Gaps = 1/127 (0%)
Frame = -3
Query: 686 PHSAPALTR*RHGRALGPPCT*AALDPRRRWY*A*SARTASRATPQAARPLRCLPRQTPP 507
P P TR GP C + DPR +T +R P RC P T P
Sbjct: 434 PRPVPTTTR-------GPVCYPGSTDPR-------CPQTTTRPVPTTTAAPRCYPGSTDP 479
Query: 506 PCWWLQPAQRSRPQPPPCTCASKPR-PRHCRAPRPTPAAAARCRLGWRTELRCCRPPSPS 330
C P RP+P ++ PR P+ P PT A RC G + RC P P+
Sbjct: 480 RC----PQTTPRPEPKCYPGSTDPRCPQTTTRPVPTTTPALRCYPG-SNDPRCPTTPRPT 534
Query: 329 SGTRRNP 309
T P
Sbjct: 535 PTTTAAP 541
Score = 42.7 bits (99), Expect = 0.007
Identities = 47/149 (31%), Positives = 54/149 (35%), Gaps = 8/149 (5%)
Frame = -3
Query: 716 RCRSRRRRARPHSAPALTR*RHGRALGPPCT*AALDPRRRWY*A*SARTASRATPQAARP 537
RC R P + PA P C + DPR T R TP P
Sbjct: 826 RCPQTTPRPVPTTTPA-----------PRCYPGSNDPR--------CPTTPRPTPTTQPP 866
Query: 536 LRCLPRQTPPPCWWLQPAQRSRPQP-----PPCTCASK-PR-PRHCRAPRPTPAAAARCR 378
LRC P T P C P RP P P C S PR P+ P PT A RC
Sbjct: 867 LRCYPGSTDPRC----PQTTPRPVPTTTPAPRCYPGSNDPRCPQTTPRPVPTTTPAPRCY 922
Query: 377 LGWRTELRCCR-PPSPSSGTRRNPHRRRP 294
G + RC + P P+ + P R P
Sbjct: 923 PG-SNDPRCPQTTPRPTQPPTQPPLRCYP 950
Score = 41.2 bits (95), Expect = 0.019
Identities = 30/92 (32%), Positives = 35/92 (37%), Gaps = 4/92 (4%)
Frame = -3
Query: 635 PPCT*AALDPRRRWY*A*SARTASRATPQAARPLRCLPRQTPPPCWWLQPAQRSRPQPPP 456
P C + DPR +T R P RC P T P C P P PP
Sbjct: 541 PRCYPGSTDPR-------CPQTTPRPVPTTTPAPRCYPGSTDPRCPQTTPRPTLPPTQPP 593
Query: 455 CTC---ASKPR-PRHCRAPRPTPAAAARCRLG 372
C ++ PR P+ P PT AA RC G
Sbjct: 594 LRCYPGSTDPRCPQTTPRPVPTTTAARRCYPG 625
Score = 40.8 bits (94), Expect = 0.025
Identities = 46/145 (31%), Positives = 53/145 (35%), Gaps = 9/145 (6%)
Frame = -3
Query: 716 RCRSRRRRARPHSAPALTR*RHGRALGPPCT*AALDPRRRWY*A*SARTASRATPQAARP 537
RC R P + PA P C DPR + A+ P P
Sbjct: 328 RCPQTTARPVPTTTPA-----------PVCYPGLTDPR-------CPKPATTTQP----P 365
Query: 536 LRCLPRQTPPPCWWLQPAQRSRPQP-----PPCTCASKPRPRHCRAPRPTP--AAAARCR 378
LRC P T P C P +RP P P C S PR PRPTP +A RC
Sbjct: 366 LRCYPGSTDPRC----PQTTTRPVPTTTPAPRCYPGSND-PRCPTTPRPTPTTTSAPRCY 420
Query: 377 LGWRTELRC--CRPPSPSSGTRRNP 309
G ++ RC P P T R P
Sbjct: 421 PG-SSDPRCPQTTTPRPVPTTTRGP 444
Score = 38.9 bits (89), Expect = 0.096
Identities = 30/95 (31%), Positives = 33/95 (34%)
Frame = -3
Query: 635 PPCT*AALDPRRRWY*A*SARTASRATPQAARPLRCLPRQTPPPCWWLQPAQRSRPQPPP 456
P C + DPR +T R TP PLRC P P C P P P
Sbjct: 144 PRCYPGSNDPR-------CPQTTPRPTPTTQPPLRCYPGSNDPRCPQTTPRPVPTTTPAP 196
Query: 455 CTCASKPRPRHCRAPRPTPAAAARCRLGWRTELRC 351
PR C P T RC G T+ RC
Sbjct: 197 VCYPGSTDPR-CPKPSTTTQPPLRCYPG-STDPRC 229
Score = 38.9 bits (89), Expect = 0.096
Identities = 36/113 (31%), Positives = 45/113 (38%), Gaps = 7/113 (6%)
Frame = -3
Query: 635 PPCT*AALDPRRRWY*A*SARTASRATPQAARPLRCLPRQTPPPCWWLQPAQRSRPQP-- 462
P C + DPR +T +R P RC P P C P RP P
Sbjct: 118 PRCYPGSTDPR-------CPQTTTRPVPTTTAAPRCYPGSNDPRC----PQTTPRPTPTT 166
Query: 461 -PPCTC---ASKPR-PRHCRAPRPTPAAAARCRLGWRTELRCCRPPSPSSGTR 318
PP C ++ PR P+ P PT A C G T+ RC P PS+ T+
Sbjct: 167 QPPLRCYPGSNDPRCPQTTPRPVPTTTPAPVCYPG-STDPRC---PKPSTTTQ 215
Score = 38.9 bits (89), Expect = 0.096
Identities = 47/174 (27%), Positives = 60/174 (34%), Gaps = 14/174 (8%)
Frame = -3
Query: 773 RPTAAGCRVCGRSPVGC-----GGRCRSRRRRARPHSAPALTR*RHGRALGPPCT*AALD 609
RPT A + P+ C RC R P + PA P C + D
Sbjct: 29 RPTTARPTPTTQPPLRCYPGSTDPRCPQTTARPVPTTTPA-----------PVCYPGSTD 77
Query: 608 PRRRWY*A*SARTASRATPQAARPLRCLPRQTPPPCWWLQPAQRSRPQPPPCTCA----- 444
PR + T PLRC P P C Q + +P P T A
Sbjct: 78 PR-----------CPKPTTTTQPPLRCYPGSNDPRC-----PQTTTTRPVPTTTAAPRCY 121
Query: 443 ---SKPR-PRHCRAPRPTPAAAARCRLGWRTELRCCRPPSPSSGTRRNPHRRRP 294
+ PR P+ P PT AA RC G + RC + + T + P R P
Sbjct: 122 PGSTDPRCPQTTTRPVPTTTAAPRCYPG-SNDPRCPQTTPRPTPTTQPPLRCYP 174
Score = 38.5 bits (88), Expect = 0.12
Identities = 36/124 (29%), Positives = 40/124 (32%), Gaps = 2/124 (1%)
Frame = -3
Query: 716 RCRSRRRRARPHSAPALTR*RHGRALGPPCT*AALDPRRRWY*A*SARTASRATPQAARP 537
RC R P + PA P C + DPR + T P
Sbjct: 778 RCPQTTPRPVPTTTPA-----------PVCYPGSTDPR-----------CPKPTTTTQPP 815
Query: 536 LRCLPRQTPPPCWWLQPAQRSRPQPPPCTCASKPRPRHCRAPRPTPAA--AARCRLGWRT 363
LRC P T P C P P P PR PRPTP RC G T
Sbjct: 816 LRCYPGSTDPRCPQTTPRPVPTTTPAPRCYPGSNDPRCPTTPRPTPTTQPPLRCYPG-ST 874
Query: 362 ELRC 351
+ RC
Sbjct: 875 DPRC 878
Score = 38.5 bits (88), Expect = 0.12
Identities = 41/132 (31%), Positives = 47/132 (35%), Gaps = 7/132 (5%)
Frame = -3
Query: 716 RCRSRRRRARPHSAPALTR*RHGRALGPPCT*AALDPRRRWY*A*SARTASRATPQAARP 537
RC RA P + PA P C + DPR + T P
Sbjct: 704 RCPQTTPRALPTTTPA-----------PVCYPGSTDPR-----------CPKPTTTTQPP 741
Query: 536 LRCLPRQTPPPCWWLQPAQRSRPQP-----PPCTCASK-PR-PRHCRAPRPTPAAAARCR 378
LRC P T P C P RP P P C S PR P+ P PT A C
Sbjct: 742 LRCYPGSTDPRC----PQTTPRPVPTTTPAPRCYPGSNDPRCPQTTPRPVPTTTPAPVCY 797
Query: 377 LGWRTELRCCRP 342
G T+ RC +P
Sbjct: 798 PG-STDPRCPKP 808
Score = 37.0 bits (84), Expect = 0.36
Identities = 30/114 (26%), Positives = 33/114 (28%), Gaps = 2/114 (1%)
Frame = -3
Query: 716 RCRSRRRRARPHSAPALTR*RHGRALGPPCT*AALDPRRRWY*A*SARTASRATPQAARP 537
RC R P + PA P C + DPR +T R P
Sbjct: 877 RCPQTTPRPVPTTTPA-----------PRCYPGSNDPR-------CPQTTPRPVPTTTPA 918
Query: 536 LRCLPRQTPPPCWWLQPAQRSRPQPPPCTCASKPRPRHC--RAPRPTPAAAARC 381
RC P P C P P PP C C PRP P C
Sbjct: 919 PRCYPGSNDPRCPQTTPRPTQPPTQPPLRCYPGSADPRCPQTTPRPVPTTRQPC 972
Score = 36.6 bits (83), Expect = 0.47
Identities = 42/152 (27%), Positives = 52/152 (33%), Gaps = 9/152 (5%)
Frame = -3
Query: 770 PTAAGCRVCGRSPVGCGGRCRSRRRRARPHSAPALTR*RHGRALGPPCT*AALDPRRRWY 591
PT R C P RC R P + PA P C + DPR
Sbjct: 614 PTTTAARRC--YPGSNDPRCPQTTPRPVPTTTPA-----------PVCYPGSTDPR---- 656
Query: 590 *A*SARTASRATPQAARPLRCLPRQTPPPCWWLQPAQRSRPQPPPCTCASK---PRPRHC 420
+ T PLRC P T P C ++ P+P P T + P
Sbjct: 657 -------CPKPTTTTQPPLRCYPGSTDPRC------PQTTPRPVPTTTPAPRCYPGSNDP 703
Query: 419 RAPRPTPAA------AARCRLGWRTELRCCRP 342
R P+ TP A A C G T+ RC +P
Sbjct: 704 RCPQTTPRALPTTTPAPVCYPG-STDPRCPKP 734
Score = 36.2 bits (82), Expect = 0.62
Identities = 40/132 (30%), Positives = 46/132 (34%), Gaps = 7/132 (5%)
Frame = -3
Query: 716 RCRSRRRRARPHSAPALTR*RHGRALGPPCT*AALDPRRRWY*A*SARTASRATPQAARP 537
RC R P + PA P C + DPR + T P
Sbjct: 254 RCPQTTPRPVPTTTPA-----------PVCYPGSTDPR-----------CPKPTTTTQPP 291
Query: 536 LRCLPRQTPPPCWWLQPAQRSRPQP-----PPCTCASK-PR-PRHCRAPRPTPAAAARCR 378
LRC P T P C P RP P P C S PR P+ P PT A C
Sbjct: 292 LRCYPGSTDPRC----PQTTPRPVPTTTPAPRCYPGSNDPRCPQTTARPVPTTTPAPVCY 347
Query: 377 LGWRTELRCCRP 342
G T+ RC +P
Sbjct: 348 PG-LTDPRCPKP 358
Score = 34.7 bits (78), Expect = 1.8
Identities = 39/132 (29%), Positives = 46/132 (34%), Gaps = 7/132 (5%)
Frame = -3
Query: 716 RCRSRRRRARPHSAPALTR*RHGRALGPPCT*AALDPRRRWY*A*SARTASRATPQAARP 537
RC R P + PA P C + DPR + + P
Sbjct: 180 RCPQTTPRPVPTTTPA-----------PVCYPGSTDPR-----------CPKPSTTTQPP 217
Query: 536 LRCLPRQTPPPCWWLQPAQRSRPQP-----PPCTCASK-PR-PRHCRAPRPTPAAAARCR 378
LRC P T P C P RP P P C S PR P+ P PT A C
Sbjct: 218 LRCYPGSTDPRC----PQTTPRPVPTTTPAPRCYPGSNDPRCPQTTPRPVPTTTPAPVCY 273
Query: 377 LGWRTELRCCRP 342
G T+ RC +P
Sbjct: 274 PG-STDPRCPKP 284
Score = 32.3 bits (72), Expect = 8.9
Identities = 34/111 (30%), Positives = 41/111 (36%), Gaps = 2/111 (1%)
Frame = -3
Query: 635 PPCT*AALDPRRRWY*A*SARTASRATPQAARPLRCLPRQTPPPCWWLQPAQRSRPQPPP 456
P C + DPR T R P R C P T P C P +RP P
Sbjct: 417 PRCYPGSSDPRC------PQTTTPRPVPTTTRGPVCYPGSTDPRC----PQTTTRP-VPT 465
Query: 455 CTCASK--PRPRHCRAPRPTPAAAARCRLGWRTELRCCRPPSPSSGTRRNP 309
T A + P R P+ TP +C G T+ RC P + TR P
Sbjct: 466 TTAAPRCYPGSTDPRCPQTTPRPEPKCYPG-STDPRC-----PQTTTRPVP 510