Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002380A_C01 KCC002380A_c01
(885 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
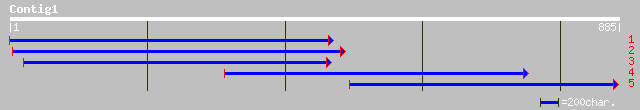
pir||T08176 glucose-1-phosphate adenylyltransferase (EC 2.7.7.27... 236 e-111
gb|AAK27718.1|AF356002_1 ADP-glucose pyrophosphorylase [Cicer ar... 168 6e-83
gb|AAM95945.1| ADP-glucose pyrophosphorylase large subunit [Onci... 164 5e-80
pir||T07674 glucose-1-phosphate adenylyltransferase (EC 2.7.7.27... 162 6e-80
ref|NP_174089.1| glucose-1-phosphate adenylyltransferase, large ... 161 8e-79
>pir||T08176 glucose-1-phosphate adenylyltransferase (EC 2.7.7.27) large chain,
precursor - Chlamydomonas reinhardtii
gi|1149717|emb|CAA62861.1| glucose-1-phosphate
adenylyltransferase [Chlamydomonas reinhardtii]
Length = 289
Score = 236 bits (603), Expect(2) = e-111
Identities = 106/107 (99%), Positives = 106/107 (99%)
Frame = +1
Query: 532 HRRRRAPVQLAAGGHQEPRHRGRAHPVGRPPVPHGLHEVRELPPRDQRRHHHRLHRLRLG 711
HRRRR PVQLAAGGHQEPRHRGRAHPVGRPPVPHGLHEVRELPPRDQRRHHHRLHRLRLG
Sbjct: 183 HRRRRGPVQLAAGGHQEPRHRGRAHPVGRPPVPHGLHEVRELPPRDQRRHHHRLHRLRLG 242
Query: 712 PRQGVRPDEDRREAPRDVVRGEAQDPGGAGRHEGGHHRAGPDPRGGC 852
PRQGVRPDEDRREAPRDVVRGEAQDPGGAGRHEGGHHRAGPDPRGGC
Sbjct: 243 PRQGVRPDEDRREAPRDVVRGEAQDPGGAGRHEGGHHRAGPDPRGGC 289
Score = 189 bits (481), Expect(2) = e-111
Identities = 90/100 (90%), Positives = 92/100 (92%)
Frame = +2
Query: 215 GPHQLGAVHHSGWRRRHPPVPADQVARQAGRAHRRRLPPDRRAHEQLHQQRHQQDLHPDP 394
GP ++GA H RHPPVPADQVARQAGRAHRRRLPPDRRAHEQLHQQRHQQDLHPDP
Sbjct: 79 GP-EVGAGDHRIQAYRHPPVPADQVARQAGRAHRRRLPPDRRAHEQLHQQRHQQDLHPDP 137
Query: 395 VQLDLPEPPPGSRLQHGQRRALRRRRLCGGAGGHPDAHRQ 514
VQLDLPEPPPGSRLQHGQRRALRRRRLCGGAGGHPDAHRQ
Sbjct: 138 VQLDLPEPPPGSRLQHGQRRALRRRRLCGGAGGHPDAHRQ 177
Score = 41.6 bits (96), Expect = 0.018
Identities = 39/114 (34%), Positives = 46/114 (40%), Gaps = 11/114 (9%)
Frame = +1
Query: 532 HRRRRAPVQLAAGGHQEPRHRGRAHPVGRPPVPHGLHEVRELPPRDQRRHHHR--LHRLR 705
HRRR P + A + RH+ HP PV L E PP + +H R L R R
Sbjct: 110 HRRRLPPDRRAHEQLHQQRHQQDLHP---DPVQLDLPEP---PPGSRLQHGQRRALRRRR 163
Query: 706 L---------GPRQGVRPDEDRREAPRDVVRGEAQDPGGAGRHEGGHHRAGPDP 840
L RQGV P RR P + G Q+P RH G H G P
Sbjct: 164 LCGGAGGHPDAHRQGVVPGHRRRRGPVQLAAGGHQEP----RHRGRAHPVGRPP 213
Score = 34.3 bits (77), Expect = 2.8
Identities = 31/105 (29%), Positives = 40/105 (37%), Gaps = 4/105 (3%)
Frame = +2
Query: 212 QGPHQLGAVHHSGWRRRHPPVPADQVARQAGRAHRRRLPP--DRRAHEQLHQQR--HQQD 379
+GP QL A H R R P + G R LPP RR H +LH+ R +Q
Sbjct: 187 RGPVQLAAGGHQEPRHRGRAHPVGRPPVPHGLHEVRELPPRDQRRHHHRLHRLRLGPRQG 246
Query: 380 LHPDPVQLDLPEPPPGSRLQHGQRRALRRRRLCGGAGGHPDAHRQ 514
+ PD + + P Q GGAG H H +
Sbjct: 247 VRPDEDRREAPRDVVRGEAQDP-----------GGAGRHEGGHHR 280
>gb|AAK27718.1|AF356002_1 ADP-glucose pyrophosphorylase [Cicer arietinum]
Length = 525
Score = 168 bits (425), Expect(2) = 6e-83
Identities = 96/177 (54%), Positives = 119/177 (66%), Gaps = 5/177 (2%)
Frame = +3
Query: 3 TLQAKSRAAASSARSEKAVPASFSSGSFRGQAVARPAATRAVRSVAARSRRAQAVKAIIE 182
T+Q + S R+ + V F +G G+ + A A+ + ++ Q + +
Sbjct: 14 TVQLREPTIMGSTRNLRFV--KFCNGEIMGRKIELKAGG-AISNCCTKNVIRQNISMSLT 70
Query: 183 APL-----LRYEPATKARTSSVLSIILGGGAGTRLFPLTKSRAKPAVPIGGAYRLIDVPM 347
A + LR K S+V++IILGGGAGTRLFPLTK RAKPAVPIGGAYRLIDVPM
Sbjct: 71 ADVAAESKLRNVDLEKRDPSTVVAIILGGGAGTRLFPLTKRRAKPAVPIGGAYRLIDVPM 130
Query: 348 SNCINSGISKIYILTQFNSTSLNRHLGRAYNMGSGVRFGGDGFVEVLAATQTPTDNG 518
SNCINSGI+K+YILTQFNS SLNRH+ RAYN G+GV F GDG+VEVLAATQTP + G
Sbjct: 131 SNCINSGINKVYILTQFNSASLNRHIARAYNSGTGVTF-GDGYVEVLAATQTPGEQG 186
Score = 162 bits (411), Expect(2) = 6e-83
Identities = 80/118 (67%), Positives = 95/118 (79%)
Frame = +2
Query: 530 GTADAVRQYSWLLEDTKNRAIEDVLILSGDHLYRMDYMKFVNYHRETNADITIGCIAYGS 709
GTADAVRQ+ WL ED +++ IEDVLILSGDHLYRMDYM FV HRE+ ADIT+ C+
Sbjct: 192 GTADAVRQFHWLFEDARSKDIEDVLILSGDHLYRMDYMDFVQNHRESGADITLSCLPMDD 251
Query: 710 DRAKEFGLMKIDEKRRVTSFAEKPKTQEALDAMKVDTTVLGLTPEEAAEKPYIASMGI 883
RA +FGLMKID K RV SF+EKPK ++ L AM+VDTTVLGL+ +EA EKPYIASMG+
Sbjct: 252 SRASDFGLMKIDNKGRVLSFSEKPKGED-LKAMQVDTTVLGLSKDEALEKPYIASMGV 308
>gb|AAM95945.1| ADP-glucose pyrophosphorylase large subunit [Oncidium cv.
'Goldiana']
Length = 517
Score = 164 bits (414), Expect(2) = 5e-80
Identities = 96/174 (55%), Positives = 115/174 (65%), Gaps = 6/174 (3%)
Frame = +3
Query: 15 KSRAAASSARSEK-AVPASFSSGSFRGQAVARPAATRAVRSVAARSRRAQ-----AVKAI 176
KS+ A S+A + A FS G GQ + + RA + R R + A +
Sbjct: 6 KSQIALSAAVLQPHAGSGRFSDGETMGQRLDFSSFRRANEQICRRRRASLLRMSFATTEL 65
Query: 177 IEAPLLRYEPATKARTSSVLSIILGGGAGTRLFPLTKSRAKPAVPIGGAYRLIDVPMSNC 356
+ LR +V+++ILGGGAGTRLFPLT+ RAKPAVPIGGAYRLIDVPMSNC
Sbjct: 66 VAEGKLRDLELEGRDPRTVVAVILGGGAGTRLFPLTRQRAKPAVPIGGAYRLIDVPMSNC 125
Query: 357 INSGISKIYILTQFNSTSLNRHLGRAYNMGSGVRFGGDGFVEVLAATQTPTDNG 518
INSGI+K+YILTQFNS SLNRHL RAYN +G+ F GDGFVEVLAATQTP + G
Sbjct: 126 INSGINKVYILTQFNSASLNRHLLRAYNFSNGIGF-GDGFVEVLAATQTPGEAG 178
Score = 157 bits (397), Expect(2) = 5e-80
Identities = 80/128 (62%), Positives = 97/128 (75%)
Frame = +2
Query: 500 DAHRQWSGSSGTADAVRQYSWLLEDTKNRAIEDVLILSGDHLYRMDYMKFVNYHRETNAD 679
+A ++W GTADAVRQ+ WL ED K + IEDVLILSGDHLYRMDYM FV HR++ AD
Sbjct: 176 EAGKKWF--QGTADAVRQFHWLFEDAKGKEIEDVLILSGDHLYRMDYMDFVQSHRQSGAD 233
Query: 680 ITIGCIAYGSDRAKEFGLMKIDEKRRVTSFAEKPKTQEALDAMKVDTTVLGLTPEEAAEK 859
ITI C+ RA +FGLMKID RV SF+EKPK QE L AM+VDT+VLGL+ E+A +
Sbjct: 234 ITISCVPMDVSRASDFGLMKIDNNGRVLSFSEKPKGQE-LKAMEVDTSVLGLSREQAKKT 292
Query: 860 PYIASMGI 883
P+IASMG+
Sbjct: 293 PFIASMGV 300
>pir||T07674 glucose-1-phosphate adenylyltransferase (EC 2.7.7.27) isoform L3
large chain - tomato gi|1840116|gb|AAC49943.1|
ADP-glucose pyrophosphorylase large subunit
[Lycopersicon esculentum]
Length = 516
Score = 162 bits (410), Expect(2) = 6e-80
Identities = 81/104 (77%), Positives = 90/104 (85%)
Frame = +3
Query: 207 ATKARTSSVLSIILGGGAGTRLFPLTKSRAKPAVPIGGAYRLIDVPMSNCINSGISKIYI 386
A K +V++IILGGG GTRLFPLTK RAKPAVPIGGAYRLIDVPMSNCINSGI+K+YI
Sbjct: 74 AKKEDARTVVAIILGGGGGTRLFPLTKRRAKPAVPIGGAYRLIDVPMSNCINSGINKVYI 133
Query: 387 LTQFNSTSLNRHLGRAYNMGSGVRFGGDGFVEVLAATQTPTDNG 518
LTQFNS SLNRH+ RAYN G+GV F GDG+VEVLAATQTP + G
Sbjct: 134 LTQFNSASLNRHIARAYNFGNGVTF-GDGYVEVLAATQTPGELG 176
Score = 158 bits (400), Expect(2) = 6e-80
Identities = 79/118 (66%), Positives = 93/118 (77%)
Frame = +2
Query: 530 GTADAVRQYSWLLEDTKNRAIEDVLILSGDHLYRMDYMKFVNYHRETNADITIGCIAYGS 709
GTADAVRQ+ WL ED +++ IEDVLILSGDHLYRMDY+ FV HR++ ADITI +
Sbjct: 182 GTADAVRQFHWLFEDARSKDIEDVLILSGDHLYRMDYLHFVQSHRQSGADITISSLPIDD 241
Query: 710 DRAKEFGLMKIDEKRRVTSFAEKPKTQEALDAMKVDTTVLGLTPEEAAEKPYIASMGI 883
RA +FGLMKID+ RV SF+EKPK + L AM VDTTVLGL+PEEA EKPYIASMG+
Sbjct: 242 SRASDFGLMKIDDTGRVMSFSEKPKGDD-LKAMAVDTTVLGLSPEEAKEKPYIASMGV 298
>ref|NP_174089.1| glucose-1-phosphate adenylyltransferase, large subunit 2,
chloroplast (ADP-glucose pyrophosphorylase) (APL2)
[Arabidopsis thaliana] gi|12644324|sp|P55230|GLG2_ARATH
Glucose-1-phosphate adenylyltransferase large subunit 2,
chloroplast precursor (ADP-glucose synthase)
(ADP-glucose pyrophosphorylase) (AGPASE S)
(Alpha-D-glucose-1-phosphate adenyl transferase)
gi|25288667|pir||G86401 protein T22C5.13 [imported] -
Arabidopsis thaliana gi|6693019|gb|AAF24945.1|AC012375_8
T22C5.13 [Arabidopsis thaliana]
gi|17380942|gb|AAL36283.1| putative ADP-glucose
pyrophosphorylase [Arabidopsis thaliana]
gi|20258949|gb|AAM14190.1| putative ADP-glucose
pyrophosphorylase [Arabidopsis thaliana]
Length = 518
Score = 161 bits (408), Expect(2) = 8e-79
Identities = 91/156 (58%), Positives = 107/156 (68%), Gaps = 10/156 (6%)
Frame = +3
Query: 81 SFRGQAVARPAATRAVRSVAARSRRAQA--VKAII--------EAPLLRYEPATKARTSS 230
+F G V +P R + +A ++ Q +++++ PLLR + A +
Sbjct: 28 AFWGTQVVKPNHLRTTKLRSAPQKKIQTNLIRSVLTPFVDQESHEPLLRTQ---NADPKN 84
Query: 231 VLSIILGGGAGTRLFPLTKSRAKPAVPIGGAYRLIDVPMSNCINSGISKIYILTQFNSTS 410
V SIILGGGAGTRLFPLT RAKPAVPIGG YRLID+PMSNCINSGI KI+ILTQFNS S
Sbjct: 85 VASIILGGGAGTRLFPLTSKRAKPAVPIGGCYRLIDIPMSNCINSGIRKIFILTQFNSFS 144
Query: 411 LNRHLGRAYNMGSGVRFGGDGFVEVLAATQTPTDNG 518
LNRHL R YN G+GV F GDGFVEVLAATQT D G
Sbjct: 145 LNRHLSRTYNFGNGVNF-GDGFVEVLAATQTSGDAG 179
Score = 155 bits (392), Expect(2) = 8e-79
Identities = 75/128 (58%), Positives = 96/128 (74%)
Frame = +2
Query: 500 DAHRQWSGSSGTADAVRQYSWLLEDTKNRAIEDVLILSGDHLYRMDYMKFVNYHRETNAD 679
DA ++W GTADAVRQ+ W+ ED K + +E VLILSGDHLYRMDYM FV H E+NAD
Sbjct: 177 DAGKKWF--QGTADAVRQFIWVFEDAKTKNVEHVLILSGDHLYRMDYMNFVQKHIESNAD 234
Query: 680 ITIGCIAYGSDRAKEFGLMKIDEKRRVTSFAEKPKTQEALDAMKVDTTVLGLTPEEAAEK 859
IT+ C+ RA +FGL+KID+ ++ F+EKPK + L AM+VDT++LGL P+EAAE
Sbjct: 235 ITVSCLPMDESRASDFGLLKIDQSGKIIQFSEKPKGDD-LKAMQVDTSILGLPPKEAAES 293
Query: 860 PYIASMGI 883
PYIASMG+
Sbjct: 294 PYIASMGV 301