Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002342A_C01 KCC002342A_c01
(564 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
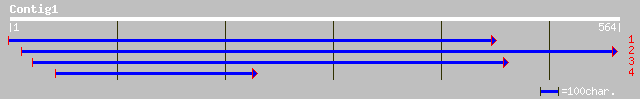
ref|XP_285023.1| similar to Retrovirus-related POL polyprotein [... 37 0.14
ref|NP_053254.1| Probable trbE gene [Agrobacterium tumefaciens] ... 34 1.6
gb|AAM61707.1| transfactor, putative [Arabidopsis thaliana] 34 1.6
ref|XP_151655.1| hypothetical protein XP_151655 [Mus musculus] 33 2.0
gb|AAM34335.1| similar to Plasmodium falciparum. Hypothetical pr... 32 5.9
>ref|XP_285023.1| similar to Retrovirus-related POL polyprotein [Mus musculus]
Length = 300
Score = 37.4 bits (85), Expect = 0.14
Identities = 26/71 (36%), Positives = 33/71 (45%)
Frame = -1
Query: 516 WRINESSNELCTDSCPMQLSTCVSRVTCAGLKLLYIFSHAAISFSAPSGLPDSRFELSTE 337
WR+ ES +LCT C T VSR T + L FS + + + LPDS E S
Sbjct: 224 WRLEESL-QLCTPECGFDRVTAVSRTTASDLWEGKGFSPGSTLYYS---LPDSELETSLA 279
Query: 336 VAEPGLDCVLP 304
P LD + P
Sbjct: 280 TTAPFLDDIQP 290
>ref|NP_053254.1| Probable trbE gene [Agrobacterium tumefaciens]
gi|6498187|dbj|BAA87639.1| tiorf14 [Agrobacterium
tumefaciens]
Length = 822
Score = 33.9 bits (76), Expect = 1.6
Identities = 22/62 (35%), Positives = 31/62 (49%)
Frame = -2
Query: 389 ASPRPVACPIADLSSQPKSQSRAWTACCQQHGCDSSTRSLAVTPSFRKVTITTNVLL*RQ 210
ASPRP CP+A+LS+ RAW A + + + + VTP +R VL+
Sbjct: 522 ASPRPAFCPLAELST---DGDRAWAA--EWIETLVALQGVTVTPDYRNAISRQLVLMAES 576
Query: 209 RG 204
RG
Sbjct: 577 RG 578
>gb|AAM61707.1| transfactor, putative [Arabidopsis thaliana]
Length = 413
Score = 33.9 bits (76), Expect = 1.6
Identities = 20/55 (36%), Positives = 26/55 (46%), Gaps = 3/55 (5%)
Frame = -1
Query: 243 HNYHQCPIIEATRRYWLSTRPSSQNNPRPLYPGAGGKYIGYLV---SCYCFVTRV 88
H YH+ P +W+S+ NNP P +GG GYL S YC V+ V
Sbjct: 26 HRYHKLP-----NSFWVSSGQELMNNPVPCQSVSGGNSGGYLFPSSSGYCNVSAV 75
>ref|XP_151655.1| hypothetical protein XP_151655 [Mus musculus]
Length = 578
Score = 33.5 bits (75), Expect = 2.0
Identities = 24/70 (34%), Positives = 35/70 (49%), Gaps = 5/70 (7%)
Frame = -1
Query: 228 CPIIEATR--RYWLSTRPSSQNNPRPLYPGAGGKYIGYLVSCYCFVTRVNLI*RLGSGLK 55
C I+ T W S + S+ PR PG G K+ G + S + V+++NL R G+
Sbjct: 366 CTIVSKTHMLEVWTSVQQDSRAGPRAWSPGTGEKFRGQVRSLHSRVSQLNLS-RFRLGIL 424
Query: 54 ENA---DLKL 34
NA DLK+
Sbjct: 425 INAPESDLKI 434
>gb|AAM34335.1| similar to Plasmodium falciparum. Hypothetical protein [Dictyostelium
discoideum]
Length = 980
Score = 32.0 bits (71), Expect = 5.9
Identities = 16/50 (32%), Positives = 22/50 (44%)
Frame = -2
Query: 392 SASPRPVACPIADLSSQPKSQSRAWTACCQQHGCDSSTRSLAVTPSFRKV 243
S SP P I+ +SS K Q CC G +S T L+ F ++
Sbjct: 859 STSPSPTTTTISPVSSPTKKQKTVNNVCCSIEGGESKTIDLSSVADFNEL 908
Database: nr
Posted date: Oct 14, 2003 12:26 AM
Number of letters in database: 498,525,298
Number of sequences in database: 1,537,769
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 546,907,673
Number of Sequences: 1537769
Number of extensions: 12001096
Number of successful extensions: 40246
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 38311
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40230
length of database: 498,525,298
effective HSP length: 117
effective length of database: 318,606,325
effective search space used: 22302442750
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)