Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002329A_C01 KCC002329A_c01
(262 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
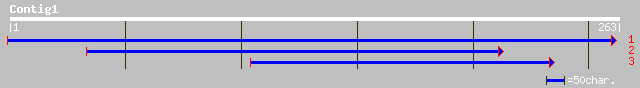
ref|NP_739198.1| putative amino acid ABC transporter glutamin-bi... 39 0.021
ref|ZP_00079120.1| COG1691: NCAIR mutase (PurE)-related proteins... 35 0.40
ref|XP_223584.2| similar to Hypothetical protein KIAA0645 [Rattu... 33 0.89
ref|XP_317169.1| ENSANGP00000018304 [Anopheles gambiae] gi|21300... 32 2.0
ref|NP_572909.1| CG15760-PA [Drosophila melanogaster] gi|2283221... 32 3.4
>ref|NP_739198.1| putative amino acid ABC transporter glutamin-binding protein
[Corynebacterium efficiens YS-314]
gi|23494431|dbj|BAC19398.1| putative amino acid ABC
transporter glutamin-binding protein [Corynebacterium
efficiens YS-314]
Length = 368
Score = 38.9 bits (89), Expect = 0.021
Identities = 24/80 (30%), Positives = 44/80 (55%), Gaps = 1/80 (1%)
Frame = +3
Query: 12 SLRGLGEGAIGVALRHLSVLKPRLRENAFSVDNQKQRT-IVVHISVHMQGGQVLAGVSPA 188
SLRGL G + + +R +SV + RLR FS + +T ++V + +QG + LAG +
Sbjct: 166 SLRGLSRGDVDMVIRSVSVTEDRLRRMEFSTPYMRTQTRLLVMDNSGIQGIEDLAGNTVC 225
Query: 189 STSPAPAAQRARSLSTRNVV 248
+ A Q+ R+++ + +
Sbjct: 226 VADGSTALQKTRAMAPESSI 245
>ref|ZP_00079120.1| COG1691: NCAIR mutase (PurE)-related proteins [Methanosarcina
barkeri]
Length = 268
Score = 34.7 bits (78), Expect = 0.40
Identities = 20/64 (31%), Positives = 33/64 (51%)
Frame = +3
Query: 45 VALRHLSVLKPRLRENAFSVDNQKQRTIVVHISVHMQGGQVLAGVSPASTSPAPAAQRAR 224
V+L HL LK ++ N + RT+VVH + + G+ A T+ PAA+ AR
Sbjct: 81 VSLGHLEALKQAFGQDKLEW-NSRARTVVVHDGTPVPRTGAVVGIVSAGTADIPAAEEAR 139
Query: 225 SLST 236
+++
Sbjct: 140 VVAS 143
>ref|XP_223584.2| similar to Hypothetical protein KIAA0645 [Rattus norvegicus]
Length = 1369
Score = 33.5 bits (75), Expect = 0.89
Identities = 20/54 (37%), Positives = 27/54 (49%), Gaps = 3/54 (5%)
Frame = +1
Query: 73 SHGFVRTPSVSTIKSNEQLLCIYRCTCKEVKFWLGFPRPAPR---RRRRPSEHG 225
S F T S T+KS + YR + + V WL FP+ APR R ++HG
Sbjct: 1293 SCSFAHTASFCTVKSYPE----YRASARAVATWLTFPQVAPRLTSRTHNGADHG 1342
>ref|XP_317169.1| ENSANGP00000018304 [Anopheles gambiae] gi|21300090|gb|EAA12235.1|
ENSANGP00000018304 [Anopheles gambiae str. PEST]
Length = 1212
Score = 32.3 bits (72), Expect = 2.0
Identities = 16/49 (32%), Positives = 25/49 (50%)
Frame = +1
Query: 10 NHCVVWEKAQSEWPSGICRFSSHGFVRTPSVSTIKSNEQLLCIYRCTCK 156
NH +W K S+WP G+ R + H + + +S KS+ L +Y K
Sbjct: 712 NHVAIWPKDASKWPKGV-RCNGHLLLNSAKMS--KSDGNFLTLYESIAK 757
>ref|NP_572909.1| CG15760-PA [Drosophila melanogaster] gi|22832212|gb|AAF48300.2|
CG15760-PA [Drosophila melanogaster]
Length = 518
Score = 31.6 bits (70), Expect = 3.4
Identities = 17/43 (39%), Positives = 19/43 (43%), Gaps = 3/43 (6%)
Frame = +2
Query: 131 CAYIGAHARRSSSGWGFPGQHLAGAG---GPASTEPQYA*CRS 250
CAY RR+ GWGF H G G PQY+ C S
Sbjct: 91 CAYSLWKKRRTLCGWGFNEHHTQSEGDSAGSCYAPPQYSRCNS 133