Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
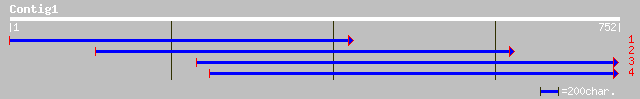
Query= KCC002163A_C01 KCC002163A_c01
(752 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAD41902.1| OSJNBa0033G05.3 [Oryza sativa (japonica cultivar... 325 6e-88
ref|NP_174005.1| E3 ubiquitin ligase SCF complex subunit Cullin,... 321 9e-87
emb|CAC85344.1| cullin 3a [Arabidopsis thaliana] 321 9e-87
emb|CAC87839.1| cullin 3B [Arabidopsis thaliana] 320 2e-86
ref|NP_177125.1| E3 ubiquitin ligase SCF complex subunit Cullin,... 320 2e-86
>emb|CAD41902.1| OSJNBa0033G05.3 [Oryza sativa (japonica cultivar-group)]
gi|32490060|emb|CAE05979.1| OSJNBa0063C18.20 [Oryza
sativa (japonica cultivar-group)]
Length = 731
Score = 325 bits (832), Expect = 6e-88
Identities = 163/222 (73%), Positives = 185/222 (82%)
Frame = +3
Query: 6 LPRELEAACESFRNFYLSTHSGRRLTFQPNMGTADLRAVFGAGRRHELNVSTYQMCVLLL 185
LP E+ E FR FYL TH+GRRLT+Q NMG AD++A FG GRRHELNVSTYQMCVL+L
Sbjct: 511 LPPEIVDISEKFRAFYLGTHNGRRLTWQTNMGNADIKATFG-GRRHELNVSTYQMCVLML 569
Query: 186 FNEADSLSYRDIAQATEIPAPDLKRALQSLACVKGRNVLRKEPAGKDVADSDVFFYNDKF 365
FN AD L+Y DI QAT IP DLKR LQSLACVKG+NVLRKEP KD+++ D F+YNDKF
Sbjct: 570 FNSADGLTYGDIEQATGIPHADLKRCLQSLACVKGKNVLRKEPMSKDISEDDTFYYNDKF 629
Query: 366 TSKLIKVKISTVAATKEGESEKAETRQKVEEDRKPQIEAAIVRIMKARQRLDHNTIITEV 545
TSKL+KVKI TV A KE E EK ETRQ+VEEDRKPQIEAAIVRIMK+R+ LDHN+IITEV
Sbjct: 630 TSKLVKVKIGTVVAQKETEPEKLETRQRVEEDRKPQIEAAIVRIMKSRRVLDHNSIITEV 689
Query: 546 TRQLQARFVPNPATIKKRIESLIEREFLARDEADRKFYTYVA 671
T+QLQ+RF+PNP IKKRIESLIEREFL RD+ DRK Y Y+A
Sbjct: 690 TKQLQSRFLPNPVVIKKRIESLIEREFLERDKVDRKMYRYLA 731
>ref|NP_174005.1| E3 ubiquitin ligase SCF complex subunit Cullin, putative [Arabidopsis
thaliana] gi|25354602|pir||A86395 hypothetical protein
T2P11.2 [imported] - Arabidopsis thaliana
gi|4262186|gb|AAD14503.1| Highly similar to cullin 3
[Arabidopsis thaliana]
gi|9295728|gb|AAF87034.1|AC006535_12 T24P13.25
[Arabidopsis thaliana] gi|34364502|emb|CAC87120.1| cullin
3a [Arabidopsis thaliana]
Length = 732
Score = 321 bits (822), Expect = 9e-87
Identities = 155/223 (69%), Positives = 184/223 (82%)
Frame = +3
Query: 3 NLPRELEAACESFRNFYLSTHSGRRLTFQPNMGTADLRAVFGAGRRHELNVSTYQMCVLL 182
NLP E+ CE FR++YL TH+GRRL++Q NMGTAD++A+FG G++HELNVST+QMCVL+
Sbjct: 510 NLPAEVSVLCEKFRSYYLGTHTGRRLSWQTNMGTADIKAIFGKGQKHELNVSTFQMCVLM 569
Query: 183 LFNEADSLSYRDIAQATEIPAPDLKRALQSLACVKGRNVLRKEPAGKDVADSDVFFYNDK 362
LFN +D LSY++I QATEIPA DLKR LQSLACVKG+NV++KEP KD+ + D+F NDK
Sbjct: 570 LFNNSDRLSYKEIEQATEIPAADLKRCLQSLACVKGKNVIKKEPMSKDIGEEDLFVVNDK 629
Query: 363 FTSKLIKVKISTVAATKEGESEKAETRQKVEEDRKPQIEAAIVRIMKARQRLDHNTIITE 542
FTSK KVKI TV A KE E EK ETRQ+VEEDRKPQIEAAIVRIMK+R+ LDHN II E
Sbjct: 630 FTSKFYKVKIGTVVAQKETEPEKQETRQRVEEDRKPQIEAAIVRIMKSRKILDHNNIIAE 689
Query: 543 VTRQLQARFVPNPATIKKRIESLIEREFLARDEADRKFYTYVA 671
VT+QLQ RF+ NP IKKRIESLIER+FL RD DRK Y Y+A
Sbjct: 690 VTKQLQPRFLANPTEIKKRIESLIERDFLERDSTDRKLYRYLA 732
>emb|CAC85344.1| cullin 3a [Arabidopsis thaliana]
Length = 338
Score = 321 bits (822), Expect = 9e-87
Identities = 155/223 (69%), Positives = 184/223 (82%)
Frame = +3
Query: 3 NLPRELEAACESFRNFYLSTHSGRRLTFQPNMGTADLRAVFGAGRRHELNVSTYQMCVLL 182
NLP E+ CE FR++YL TH+GRRL++Q NMGTAD++A+FG G++HELNVST+QMCVL+
Sbjct: 116 NLPAEVSVLCEKFRSYYLGTHTGRRLSWQTNMGTADIKAIFGKGQKHELNVSTFQMCVLM 175
Query: 183 LFNEADSLSYRDIAQATEIPAPDLKRALQSLACVKGRNVLRKEPAGKDVADSDVFFYNDK 362
LFN +D LSY++I QATEIPA DLKR LQSLACVKG+NV++KEP KD+ + D+F NDK
Sbjct: 176 LFNNSDRLSYKEIEQATEIPAADLKRCLQSLACVKGKNVIKKEPMSKDIGEEDLFVVNDK 235
Query: 363 FTSKLIKVKISTVAATKEGESEKAETRQKVEEDRKPQIEAAIVRIMKARQRLDHNTIITE 542
FTSK KVKI TV A KE E EK ETRQ+VEEDRKPQIEAAIVRIMK+R+ LDHN II E
Sbjct: 236 FTSKFYKVKIGTVVAQKETEPEKQETRQRVEEDRKPQIEAAIVRIMKSRKILDHNNIIAE 295
Query: 543 VTRQLQARFVPNPATIKKRIESLIEREFLARDEADRKFYTYVA 671
VT+QLQ RF+ NP IKKRIESLIER+FL RD DRK Y Y+A
Sbjct: 296 VTKQLQPRFLANPTEIKKRIESLIERDFLERDSTDRKLYRYLA 338
>emb|CAC87839.1| cullin 3B [Arabidopsis thaliana]
Length = 601
Score = 320 bits (819), Expect = 2e-86
Identities = 155/223 (69%), Positives = 183/223 (81%)
Frame = +3
Query: 3 NLPRELEAACESFRNFYLSTHSGRRLTFQPNMGTADLRAVFGAGRRHELNVSTYQMCVLL 182
NLP E+ CE FR++YL TH+GRRL++Q NMGTAD++AVFG G++HELNVST+QMCVL+
Sbjct: 379 NLPAEVSVLCEKFRSYYLGTHTGRRLSWQTNMGTADIKAVFGKGQKHELNVSTFQMCVLM 438
Query: 183 LFNEADSLSYRDIAQATEIPAPDLKRALQSLACVKGRNVLRKEPAGKDVADSDVFFYNDK 362
LFN +D LSY++I QATEIP PDLKR LQS+ACVKG+NVLRKEP K++A+ D F ND+
Sbjct: 439 LFNNSDRLSYKEIEQATEIPTPDLKRCLQSMACVKGKNVLRKEPMSKEIAEEDWFVVNDR 498
Query: 363 FTSKLIKVKISTVAATKEGESEKAETRQKVEEDRKPQIEAAIVRIMKARQRLDHNTIITE 542
F SK KVKI TV A KE E EK ETRQ+VEEDRKPQIEAAIVRIMK+R+ LDHN II E
Sbjct: 499 FASKFYKVKIGTVVAQKETEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRVLDHNNIIAE 558
Query: 543 VTRQLQARFVPNPATIKKRIESLIEREFLARDEADRKFYTYVA 671
VT+QLQ RF+ NP IKKRIESLIER+FL RD DRK Y Y+A
Sbjct: 559 VTKQLQTRFLANPTEIKKRIESLIERDFLERDNTDRKLYRYLA 601
>ref|NP_177125.1| E3 ubiquitin ligase SCF complex subunit Cullin, putative [Arabidopsis
thaliana] gi|25354600|pir||E96718 probable cullin
T6C23.13 [imported] - Arabidopsis thaliana
gi|12325193|gb|AAG52544.1|AC013289_11 putative cullin;
66460-68733 [Arabidopsis thaliana]
Length = 732
Score = 320 bits (819), Expect = 2e-86
Identities = 155/223 (69%), Positives = 183/223 (81%)
Frame = +3
Query: 3 NLPRELEAACESFRNFYLSTHSGRRLTFQPNMGTADLRAVFGAGRRHELNVSTYQMCVLL 182
NLP E+ CE FR++YL TH+GRRL++Q NMGTAD++AVFG G++HELNVST+QMCVL+
Sbjct: 510 NLPAEVSVLCEKFRSYYLGTHTGRRLSWQTNMGTADIKAVFGKGQKHELNVSTFQMCVLM 569
Query: 183 LFNEADSLSYRDIAQATEIPAPDLKRALQSLACVKGRNVLRKEPAGKDVADSDVFFYNDK 362
LFN +D LSY++I QATEIP PDLKR LQS+ACVKG+NVLRKEP K++A+ D F ND+
Sbjct: 570 LFNNSDRLSYKEIEQATEIPTPDLKRCLQSMACVKGKNVLRKEPMSKEIAEEDWFVVNDR 629
Query: 363 FTSKLIKVKISTVAATKEGESEKAETRQKVEEDRKPQIEAAIVRIMKARQRLDHNTIITE 542
F SK KVKI TV A KE E EK ETRQ+VEEDRKPQIEAAIVRIMK+R+ LDHN II E
Sbjct: 630 FASKFYKVKIGTVVAQKETEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRVLDHNNIIAE 689
Query: 543 VTRQLQARFVPNPATIKKRIESLIEREFLARDEADRKFYTYVA 671
VT+QLQ RF+ NP IKKRIESLIER+FL RD DRK Y Y+A
Sbjct: 690 VTKQLQTRFLANPTEIKKRIESLIERDFLERDNTDRKLYRYLA 732