Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002125A_C01 KCC002125A_c01
(868 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
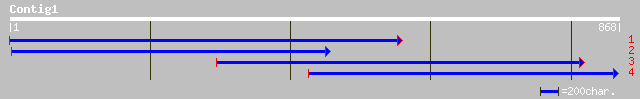
ref|NP_565735.1| CCR4-associated factor -related [Arabidopsis th... 304 1e-81
ref|NP_178193.1| CCR4-associated factor -related\0CCR4-associate... 302 4e-81
pir||F84728 probable CCR4-associated factor [imported] - Arabido... 281 8e-75
ref|NP_196657.1| CCR4-ASSOCIATED FACTOR -related protein [Arabid... 278 8e-74
gb|AAK92792.1| putative CCR4-associated factor [Arabidopsis thal... 278 8e-74
>ref|NP_565735.1| CCR4-associated factor -related [Arabidopsis thaliana]
gi|15293025|gb|AAK93623.1| putative CCR4-associated
factor [Arabidopsis thaliana] gi|20197620|gb|AAD15397.2|
putative CCR4-associated factor [Arabidopsis thaliana]
gi|23296713|gb|AAN13153.1| putative CCR4-associated
factor [Arabidopsis thaliana]
Length = 275
Score = 304 bits (778), Expect = 1e-81
Identities = 134/220 (60%), Positives = 183/220 (82%), Gaps = 1/220 (0%)
Frame = +3
Query: 210 DTLRVREVWADNMEVEFALLRDIVEDYPYIAMDTEFPGVVAKPMGTFKSSREYLYKALKM 389
D++++REVW DN+E E AL+R++V+D+P++AMDTEFPG+V +P+GTFK++ EY Y+ LK
Sbjct: 8 DSIQIREVWNDNLESEMALIREVVDDFPFVAMDTEFPGIVCRPVGTFKTNTEYHYETLKT 67
Query: 390 NVDMLKLIQLGLTLTDAKGTLPRAA-NGELCVWQFNFKGFKLSDDVYAQDSIELLKQSGI 566
NV++LK+IQLGLT +D KG LP + + C+WQFNF+ F L D+YA DSIELL+QSGI
Sbjct: 68 NVNILKMIQLGLTFSDEKGNLPTCGTDNKYCIWQFNFREFDLESDIYATDSIELLRQSGI 127
Query: 567 DFALHEARGIDVHRFGELLMTSGIVLNDDVRWITFHSNYDFGYLLKILTCQPLPGTEQEF 746
DF + GID RF ELLM+SGIVLN++V W+TFHS YDFGYLLK+LTCQ LP T+ F
Sbjct: 128 DFVKNNEFGIDSKRFAELLMSSGIVLNENVHWVTFHSGYDFGYLLKLLTCQNLPETQTGF 187
Query: 747 FELLNIYFPNIFDIKYLMRYCDNLHGGLNKLAEMLDVQRI 866
FE++++YFP ++DIK+LM++C++LHGGLNKLAE+LDV+R+
Sbjct: 188 FEMISVYFPRVYDIKHLMKFCNSLHGGLNKLAELLDVERV 227
>ref|NP_178193.1| CCR4-associated factor -related\0CCR4-associated factor, putative
[Arabidopsis thaliana] gi|30699538|ref|NP_849915.1|
CCR4-associated factor -related\0CCR4-associated factor,
putative [Arabidopsis thaliana] gi|25363292|pir||D96840
hypothetical protein F23A5.13 [imported] - Arabidopsis
thaliana gi|6503290|gb|AAF14666.1|AC011713_14 Similar to
gb|U21855 CCR4-associated factor 1 (CAF1) from Mus
musculus. ESTs gb|AAA394972, gb|AA585812 and gb|H77015
come from this gene. [Arabidopsis thaliana]
gi|17979381|gb|AAL49916.1| putative CCR4-associated
factorCCR4-associated factor [Arabidopsis thaliana]
gi|20465785|gb|AAM20381.1| putative CCR4-associated
factor [Arabidopsis thaliana]
Length = 274
Score = 302 bits (774), Expect = 4e-81
Identities = 131/219 (59%), Positives = 182/219 (82%)
Frame = +3
Query: 210 DTLRVREVWADNMEVEFALLRDIVEDYPYIAMDTEFPGVVAKPMGTFKSSREYLYKALKM 389
D++++REVW DN++ E L+RD+V+D+PY+AMDTEFPG+V +P+GTFKS+ +Y Y+ LK
Sbjct: 8 DSIQIREVWNDNLQEEMDLIRDVVDDFPYVAMDTEFPGIVVRPVGTFKSNADYHYETLKT 67
Query: 390 NVDMLKLIQLGLTLTDAKGTLPRAANGELCVWQFNFKGFKLSDDVYAQDSIELLKQSGID 569
NV++LK+IQLGLT ++ +G LP + C+WQFNF+ F L D++A DSIELLKQSGID
Sbjct: 68 NVNILKMIQLGLTFSNEQGNLPTCGTDKYCIWQFNFREFDLDSDIFALDSIELLKQSGID 127
Query: 570 FALHEARGIDVHRFGELLMTSGIVLNDDVRWITFHSNYDFGYLLKILTCQPLPGTEQEFF 749
A + GID RF ELLM+SGIVLN++V W+TFHS YDFGYLLK+LTCQ LP ++ +FF
Sbjct: 128 LAKNTLDGIDSKRFAELLMSSGIVLNENVHWVTFHSGYDFGYLLKLLTCQNLPDSQTDFF 187
Query: 750 ELLNIYFPNIFDIKYLMRYCDNLHGGLNKLAEMLDVQRI 866
+L+N+YFP ++DIK+LM++C++LHGGLNKLAE+L+V+R+
Sbjct: 188 KLINVYFPTVYDIKHLMKFCNSLHGGLNKLAELLEVERV 226
>pir||F84728 probable CCR4-associated factor [imported] - Arabidopsis thaliana
Length = 252
Score = 281 bits (720), Expect = 8e-75
Identities = 125/203 (61%), Positives = 169/203 (82%), Gaps = 1/203 (0%)
Frame = +3
Query: 261 ALLRDIVEDYPYIAMDTEFPGVVAKPMGTFKSSREYLYKALKMNVDMLKLIQLGLTLTDA 440
AL+R++V+D+P++AMDTEFPG+V +P+GTFK++ EY Y+ LK NV++LK+IQLGLT +D
Sbjct: 2 ALIREVVDDFPFVAMDTEFPGIVCRPVGTFKTNTEYHYETLKTNVNILKMIQLGLTFSDE 61
Query: 441 KGTLPRAA-NGELCVWQFNFKGFKLSDDVYAQDSIELLKQSGIDFALHEARGIDVHRFGE 617
KG LP + + C+WQFNF+ F L D+YA DSIELL+QSGIDF + GID RF E
Sbjct: 62 KGNLPTCGTDNKYCIWQFNFREFDLESDIYATDSIELLRQSGIDFVKNNEFGIDSKRFAE 121
Query: 618 LLMTSGIVLNDDVRWITFHSNYDFGYLLKILTCQPLPGTEQEFFELLNIYFPNIFDIKYL 797
LLM+SGIVLN++V W+TFHS YDFGYLLK+LTCQ LP T+ FFE++++YFP ++DIK+L
Sbjct: 122 LLMSSGIVLNENVHWVTFHSGYDFGYLLKLLTCQNLPETQTGFFEMISVYFPRVYDIKHL 181
Query: 798 MRYCDNLHGGLNKLAEMLDVQRI 866
M++C++LHGGLNKLAE+LDV+R+
Sbjct: 182 MKFCNSLHGGLNKLAELLDVERV 204
>ref|NP_196657.1| CCR4-ASSOCIATED FACTOR -related protein [Arabidopsis thaliana]
gi|11288398|pir||T50805 CCR4-ASSOCIATED FACTOR-like
protein - Arabidopsis thaliana
gi|8979730|emb|CAB96851.1| CCR4-ASSOCIATED FACTOR-like
protein [Arabidopsis thaliana]
gi|23296319|gb|AAN13040.1| putative CCR4-associated
factor [Arabidopsis thaliana]
Length = 277
Score = 278 bits (711), Expect = 8e-74
Identities = 124/219 (56%), Positives = 170/219 (77%)
Frame = +3
Query: 210 DTLRVREVWADNMEVEFALLRDIVEDYPYIAMDTEFPGVVAKPMGTFKSSREYLYKALKM 389
D++ +REVW N+ EFAL+R+IV+ + YIAMDTEFPGVV KP+ TFK + + Y+ LK
Sbjct: 8 DSIMIREVWDYNLVEEFALIREIVDKFSYIAMDTEFPGVVLKPVATFKYNNDLNYRTLKE 67
Query: 390 NVDMLKLIQLGLTLTDAKGTLPRAANGELCVWQFNFKGFKLSDDVYAQDSIELLKQSGID 569
NVD+LKLIQ+GLT +D G LP + C+WQFNF+ F + +D+YA +SIELL+Q GID
Sbjct: 68 NVDLLKLIQVGLTFSDENGNLPTCGTDKFCIWQFNFREFNIGEDIYASESIELLRQCGID 127
Query: 570 FALHEARGIDVHRFGELLMTSGIVLNDDVRWITFHSNYDFGYLLKILTCQPLPGTEQEFF 749
F + +GIDV RFGEL+M+SGIVLND + W+TFH YDFGYL+K+LTC+ LP + +FF
Sbjct: 128 FKKNIEKGIDVVRFGELMMSSGIVLNDAISWVTFHGGYDFGYLVKLLTCKELPLKQADFF 187
Query: 750 ELLNIYFPNIFDIKYLMRYCDNLHGGLNKLAEMLDVQRI 866
+LL +YFP ++DIK+LM +C+ L GGLN+LAE++ V+R+
Sbjct: 188 KLLYVYFPTVYDIKHLMTFCNGLFGGLNRLAELMGVERV 226
>gb|AAK92792.1| putative CCR4-associated factor [Arabidopsis thaliana]
Length = 277
Score = 278 bits (711), Expect = 8e-74
Identities = 124/219 (56%), Positives = 170/219 (77%)
Frame = +3
Query: 210 DTLRVREVWADNMEVEFALLRDIVEDYPYIAMDTEFPGVVAKPMGTFKSSREYLYKALKM 389
D++ +REVW N+ EFAL+R+IV+ + YIAMDTEFPGVV KP+ TFK + + Y+ LK
Sbjct: 8 DSIMIREVWDYNLVEEFALIREIVDKFSYIAMDTEFPGVVLKPVATFKYNNDLNYRTLKE 67
Query: 390 NVDMLKLIQLGLTLTDAKGTLPRAANGELCVWQFNFKGFKLSDDVYAQDSIELLKQSGID 569
NVD+LKLIQ+GLT +D G LP + C+WQFNF+ F + +D+YA +SIELL+Q GID
Sbjct: 68 NVDLLKLIQVGLTFSDENGNLPTCGTDKFCIWQFNFREFNIGEDIYASESIELLRQCGID 127
Query: 570 FALHEARGIDVHRFGELLMTSGIVLNDDVRWITFHSNYDFGYLLKILTCQPLPGTEQEFF 749
F + +GIDV RFGEL+M+SGIVLND + W+TFH YDFGYL+K+LTC+ LP + +FF
Sbjct: 128 FKKNIEKGIDVVRFGELMMSSGIVLNDAISWVTFHGGYDFGYLVKLLTCKELPLKQADFF 187
Query: 750 ELLNIYFPNIFDIKYLMRYCDNLHGGLNKLAEMLDVQRI 866
+LL +YFP ++DIK+LM +C+ L GGLN+LAE++ V+R+
Sbjct: 188 KLLYVYFPTVYDIKHLMTFCNGLFGGLNRLAELMGVERV 226