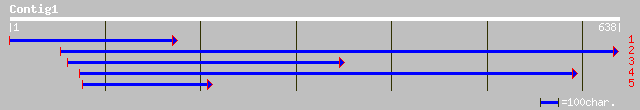
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC002005A_C01 KCC002005A_c01
(621 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO73245.1| putative membrane protein [Oryza sativa (japonica... 114 1e-24
ref|NP_173069.1| expressed protein [Arabidopsis thaliana] gi|253... 108 6e-23
gb|AAL36053.1| AT3g24470/MXP5_4 [Arabidopsis thaliana] gi|372019... 66 5e-10
ref|NP_849373.1| expressed protein [Arabidopsis thaliana] 61 1e-08
ref|NP_567403.1| expressed protein [Arabidopsis thaliana] gi|166... 61 1e-08
>gb|AAO73245.1| putative membrane protein [Oryza sativa (japonica cultivar-group)]
Length = 417
Score = 114 bits (285), Expect = 1e-24
Identities = 64/151 (42%), Positives = 96/151 (63%), Gaps = 1/151 (0%)
Frame = +3
Query: 171 YLGSCAAQLATYGSLHGPASAHLGQVLRHSARVAWSVLFFLAMILAWVLRDFATPILEKI 350
+ SC A + + ASA L + R SAR+A+ LF ++IL++++R FATP+L++I
Sbjct: 2 WCASCLASACAGCTCNLCASA-LSAISRRSARLAYCGLFAASLILSFLMRQFATPLLKQI 60
Query: 351 PWI-VKDVTQVDMDKWFGQQAVYRVSMGNFLFFGCMSLALLGVKQRGDKRDAYLHHGHPL 527
PWI D TQ D +WF AV RVS+GNFLFF +L ++GVK + D+RDA+ HHG +
Sbjct: 61 PWINTFDYTQPD--EWFQMNAVLRVSLGNFLFFAIFALMMIGVKDQNDRRDAW-HHGGWI 117
Query: 528 AKLGLWLLFHCLPFLFPNEVLNVYSWAARVG 620
AK+ +W++ L F PN V+ +Y ++ G
Sbjct: 118 AKIVVWVVLIVLMFCVPNVVITIYEVLSKFG 148
>ref|NP_173069.1| expressed protein [Arabidopsis thaliana] gi|25354033|pir||F86296
hypothetical protein T24D18.26 - Arabidopsis thaliana
gi|6587821|gb|AAF18512.1|AC010924_25 Contains similarity
to gb|AF181686 membrane protein TMS1d from Drosophila
melanogaster. ESTs gb|R64994, gb|AI994832, gb|Z47674
come from this gene. [Arabidopsis thaliana]
Length = 412
Score = 108 bits (270), Expect = 6e-23
Identities = 58/155 (37%), Positives = 87/155 (55%)
Frame = +3
Query: 156 MFLGGYLGSCAAQLATYGSLHGPASAHLGQVLRHSARVAWSVLFFLAMILAWVLRDFATP 335
MF L SC A A + + R SAR+A+ LF L++I++W+LR+ A P
Sbjct: 1 MFAASCLASCCAACAC-----DACRTVVSGISRRSARIAYCGLFALSLIVSWILREVAAP 55
Query: 336 ILEKIPWIVKDVTQVDMDKWFGQQAVYRVSMGNFLFFGCMSLALLGVKQRGDKRDAYLHH 515
++EK+PWI D + WF AV RVS+GNFLFF +S+ ++GVK + D RD +HH
Sbjct: 56 LMEKLPWINHFHKTPDRE-WFETDAVLRVSLGNFLFFSILSVMMIGVKNQKDPRDG-IHH 113
Query: 516 GHPLAKLGLWLLFHCLPFLFPNEVLNVYSWAARVG 620
G + K+ W + F PNE+++ Y ++ G
Sbjct: 114 GGWMMKIICWCILVIFMFFLPNEIISFYESMSKFG 148
>gb|AAL36053.1| AT3g24470/MXP5_4 [Arabidopsis thaliana] gi|37201996|gb|AAQ89613.1|
At3g24470/MXP5_4 [Arabidopsis thaliana]
Length = 409
Score = 65.9 bits (159), Expect = 5e-10
Identities = 38/120 (31%), Positives = 59/120 (48%)
Frame = +3
Query: 261 ARVAWSVLFFLAMILAWVLRDFATPILEKIPWIVKDVTQVDMDKWFGQQAVYRVSMGNFL 440
AR + ++F +A +LAW RD+ L K+ + + G V RVS+G FL
Sbjct: 40 ARYVYGLIFLIANLLAWAARDYGRGALRKV---TRFKNCKGGENCLGTDGVLRVSLGCFL 96
Query: 441 FFGCMSLALLGVKQRGDKRDAYLHHGHPLAKLGLWLLFHCLPFLFPNEVLNVYSWAARVG 620
F+ M L+ LG + RD + H G KL +W +PFL P+ ++++Y A G
Sbjct: 97 FYFVMFLSTLGTSKTHSSRDRW-HSGWWFVKLIMWPALTIIPFLLPSSIIHLYGEIAHFG 155
>ref|NP_849373.1| expressed protein [Arabidopsis thaliana]
Length = 394
Score = 61.2 bits (147), Expect = 1e-08
Identities = 40/120 (33%), Positives = 58/120 (48%)
Frame = +3
Query: 261 ARVAWSVLFFLAMILAWVLRDFATPILEKIPWIVKDVTQVDMDKWFGQQAVYRVSMGNFL 440
AR + ++F LA +LAW LRD+ L ++ K + G + V RVS G FL
Sbjct: 33 ARYVYGLIFLLANLLAWALRDYGRGALTEMR---KFKNCKEGGDCLGTEGVLRVSFGCFL 89
Query: 441 FFGCMSLALLGVKQRGDKRDAYLHHGHPLAKLGLWLLFHCLPFLFPNEVLNVYSWAARVG 620
F+ M L+ +G + RD + H G AKL + L PFL P+ ++ Y A G
Sbjct: 90 FYFIMFLSTVGTSKTHSSRDKW-HSGWWFAKLFMLLGLTIFPFLLPSSIIQFYGEIAHFG 148
>ref|NP_567403.1| expressed protein [Arabidopsis thaliana] gi|16604677|gb|AAL24131.1|
unknown protein [Arabidopsis thaliana]
gi|21436351|gb|AAM51345.1| unknown protein [Arabidopsis
thaliana]
Length = 394
Score = 61.2 bits (147), Expect = 1e-08
Identities = 40/120 (33%), Positives = 58/120 (48%)
Frame = +3
Query: 261 ARVAWSVLFFLAMILAWVLRDFATPILEKIPWIVKDVTQVDMDKWFGQQAVYRVSMGNFL 440
AR + ++F LA +LAW LRD+ L ++ K + G + V RVS G FL
Sbjct: 33 ARYVYGLIFLLANLLAWALRDYGRGALTEMR---KFKNCKEGGDCLGTEGVLRVSFGCFL 89
Query: 441 FFGCMSLALLGVKQRGDKRDAYLHHGHPLAKLGLWLLFHCLPFLFPNEVLNVYSWAARVG 620
F+ M L+ +G + RD + H G AKL + L PFL P+ ++ Y A G
Sbjct: 90 FYFIMFLSTVGTSKTHSSRDKW-HSGWWFAKLFMLLGLTIFPFLLPSSIIQFYGEIAHFG 148