Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001987A_C01 KCC001987A_c01
(1447 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
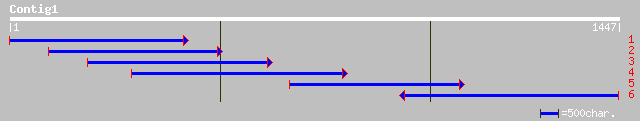
dbj|BAC04906.1| unnamed protein product [Homo sapiens] 41 0.061
dbj|BAB44032.1| P0684B02.19 [Oryza sativa (japonica cultivar-gro... 39 0.18
ref|NP_723663.1| CG32830-PA [Drosophila melanogaster] gi|2294624... 39 0.23
ref|XP_215259.2| similar to mKIAA0595 protein [Rattus norvegicus] 38 0.40
ref|ZP_00127177.1| COG3064: Membrane protein involved in colicin... 38 0.52
>dbj|BAC04906.1| unnamed protein product [Homo sapiens]
Length = 226
Score = 40.8 bits (94), Expect = 0.061
Identities = 36/143 (25%), Positives = 55/143 (38%), Gaps = 5/143 (3%)
Frame = -2
Query: 1350 THSAAHTRTHMHTHNQSPQARVSNVHRTGLPVPY*LGDMRRHRTRRLAHHYFVRRRVSAA 1171
TH+ H+ TH+H+H S + H L + H H S
Sbjct: 87 THAHMHSHTHIHSHTHSHIHSNTPTHTHPLTSTHIQTSTVTHTHSHTHAHKHTYTHTSTH 146
Query: 1170 TVRVCLSAVCERTR*L----IQCGSHSILHSRSCGATYTHAHT-PLPHTPMHWQPHESSR 1006
T + + T L +H++ HS + T+ H+HT PL HT H PH +
Sbjct: 147 THPLTFTHTYPITHPLTHTPTHTHTHALTHSHTHIHTHMHSHTHPLTHTSTHTHPHTHNV 206
Query: 1005 PDPQNDRRHMLCKIYLLSAEPCR 937
Q+ L + ++ A PCR
Sbjct: 207 WLTQD-----LVFMLMILAHPCR 224
>dbj|BAB44032.1| P0684B02.19 [Oryza sativa (japonica cultivar-group)]
gi|19571085|dbj|BAB86510.1| P0692C11.6 [Oryza sativa
(japonica cultivar-group)]
Length = 779
Score = 39.3 bits (90), Expect = 0.18
Identities = 52/225 (23%), Positives = 77/225 (34%), Gaps = 5/225 (2%)
Frame = -1
Query: 1042 HSNALATPRIKPPRPTE*QTPHAVQDIPTIRRAMPLLFCHLHVRSKCGFSTQ---P*STG 872
H N +T +PP P +P + + R P +L V G + ++
Sbjct: 521 HRNHPSTVHPEPPPPYAASSPPCSTSVTVVHRRKPTGTWYLSVAGPAGPRPRLRHGRASS 580
Query: 871 SRQSTTNVRAPPLPGAPRSLSWWPRAGKLTPARVLLFVYGSS*PFLLFWLLLRRATKPQL 692
R+S PP P S W + + R + PF LF RA P +
Sbjct: 581 DRRSRRLPCRPPPSFPPPVASPWSSRRRRSSVRTPFISF----PFPLFSSPPVRAVAPVV 636
Query: 691 PP--Y*CVSRRQRPHSRSLFPAMAVPPTPAPRSAAKPAARVSRGHPSGWLSAYRAPSEAP 518
+ V RR RP R P+ PR V R S + + P
Sbjct: 637 VAGDHRAVRRRSRPADRR--------PSWPPR-----VCHVVRPRTSACVDSVHGAPATP 683
Query: 517 DKPRSQRPLSTASNGNRGFWVGHVSRGRTRNPQAHMSIEFLNLAR 383
P RPL T ++G W+ S GR + + + + AR
Sbjct: 684 CNPCELRPLGTDTSGRNPRWIRSFSTGRPAHHSSTVGVHACRHAR 728
>ref|NP_723663.1| CG32830-PA [Drosophila melanogaster] gi|22946244|gb|AAN10775.1|
CG32830-PA [Drosophila melanogaster]
Length = 416
Score = 38.9 bits (89), Expect = 0.23
Identities = 20/60 (33%), Positives = 30/60 (49%), Gaps = 3/60 (5%)
Frame = -1
Query: 217 MQAEKGQDRK---QRGTQHRHHCRPDPTKHPRHHKHPQAHGPIARNTRSSGAHNCTPTPT 47
MQ ++ Q R+ QR + H+ H P H HH H Q NT+ + + N +PTP+
Sbjct: 93 MQQQEQQQRQLFDQRRSHHQPHHHHQPHHHRHHHHHQQRINNNNNNTKKNRSANSSPTPS 152
>ref|XP_215259.2| similar to mKIAA0595 protein [Rattus norvegicus]
Length = 1728
Score = 38.1 bits (87), Expect = 0.40
Identities = 34/107 (31%), Positives = 39/107 (35%), Gaps = 12/107 (11%)
Frame = -1
Query: 691 PPY*CVSRRQRPHSRSLFPAMAVPPTP--APRSAAKPAARVSRGHPSGWLSAYRAPSEAP 518
PP C + Q P A P P AP + P A R + R PSE P
Sbjct: 1411 PPLPCTTTSQEPLDHRTSSEQADPSAPCFAPSTLLSPEASPCRNEMNT-----RTPSEPP 1465
Query: 517 DKPRSQR----------PLSTASNGNRGFWVGHVSRGRTRNPQAHMS 407
DK RS R P S G RG VS G +R +A S
Sbjct: 1466 DKQRSMRCYRKACRSASPPSRGWQGRRGRSSRSVSSGSSRTSEASSS 1512
>ref|ZP_00127177.1| COG3064: Membrane protein involved in colicin uptake [Pseudomonas
syringae pv. syringae B728a]
Length = 133
Score = 37.7 bits (86), Expect = 0.52
Identities = 26/82 (31%), Positives = 40/82 (48%), Gaps = 7/82 (8%)
Frame = -1
Query: 718 LRRATKPQLPPY*CVSR----RQRPHSRSL---FPAMAVPPTPAPRSAAKPAARVSRGHP 560
+R++TKP+LP +P ++S PA P PAP++AAKPA + P
Sbjct: 49 VRKSTKPKLPEVDSAKAPAKGAAKPAAKSAPAPKPAGEAKPKPAPKAAAKPAPKAK---P 105
Query: 559 SGWLSAYRAPSEAPDKPRSQRP 494
+ + EAP KP ++P
Sbjct: 106 AAAAATDAPAKEAPAKPAPKKP 127