Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001795A_C01 KCC001795A_c01
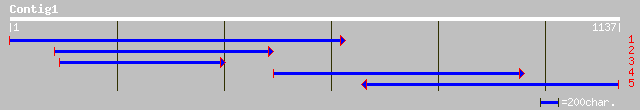
(967 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN07178.1| Crip-31 [Clerodendrum inerme] 37 0.38
ref|ZP_00082169.1| COG0741: Soluble lytic murein transglycosylas... 37 0.50
ref|ZP_00041521.1| COG2386: ABC-type transport system involved i... 35 1.5
ref|XP_330608.1| predicted protein [Neurospora crassa] gi|289263... 35 1.5
emb|CAD71233.1| hypothetical protein [Neurospora crassa] 35 1.9
>gb|AAN07178.1| Crip-31 [Clerodendrum inerme]
Length = 290
Score = 37.4 bits (85), Expect = 0.38
Identities = 37/117 (31%), Positives = 47/117 (39%)
Frame = +2
Query: 476 PACAADRQARSGCVPSWPISAAGGPRRILRPQVYTGILKIPSLSSTGQFRWSSAPSSHSH 655
PAC + + S C PS P S A PRR RP SS+ RW P+S S
Sbjct: 171 PACRSSSASSSSCTPSSPRSPACSPRR--RPS-----------SSSWPSRWPRPPASTSR 217
Query: 656 *MSSPVHPRARDCTGLLLWIRPGSDYLGLLGSSFVTSRHPMLSPKSTGQRQRRAAAS 826
P +R T RPG +R P SP++ R RR++AS
Sbjct: 218 -TYRPTSTASR--TTRCSTSRPG-------------ARSPPRSPRTACSRSRRSSAS 258
>ref|ZP_00082169.1| COG0741: Soluble lytic murein transglycosylase and related
regulatory proteins (some contain LysM/invasin domains)
[Geobacter metallireducens]
Length = 259
Score = 37.0 bits (84), Expect = 0.50
Identities = 33/120 (27%), Positives = 45/120 (37%), Gaps = 14/120 (11%)
Frame = -1
Query: 703 QPGAVACAR---------MHRAAHSMRMR*WSGGPTKLARGAERWYFQYARINLRPEDPA 551
QPGA A A M R+A ++ G P + R E +A + P DPA
Sbjct: 48 QPGARASAAAAAELMRLDMMRSAFTLGDSEGGGNPPAMGRAVEFMLKSFAENSREPVDPA 107
Query: 550 GP---AGRRDGP*RHTPRSSLPVCGTCGWSSSVLLRIDEYRGVSV--VAMWMRSEGNSTP 386
P G P LP G+ W V+ R GV V + +++E N P
Sbjct: 108 SPVAAGGSSAAPVASAAPEPLPAKGSGNWLDDVVNRASRRHGVEVGLIKAVIKAESNFNP 167
>ref|ZP_00041521.1| COG2386: ABC-type transport system involved in cytochrome c
biogenesis, permease component [Xylella fastidiosa
Ann-1]
Length = 294
Score = 35.4 bits (80), Expect = 1.5
Identities = 35/94 (37%), Positives = 42/94 (44%)
Frame = +1
Query: 268 GRTLWSCPSVGSHECKHQPLETPVRARS*PRRSPSHARESVSNSPHSASTSPPHSLLCIH 447
GRTL S G H + P TP S PRR +H +PH+AS PP SL
Sbjct: 11 GRTLRQPRSRGHHTPQPHPHRTP----SHPRRHTAH---HTRRAPHTASPHPPASLTESS 63
Query: 448 QSAIGPSLTTRMCRRQASSIGVCAFMAHLCGRRA 549
SA P L+ QASS + + L RRA
Sbjct: 64 MSAPIPPLSL----WQASSAIITRDLRLLWRRRA 93
>ref|XP_330608.1| predicted protein [Neurospora crassa] gi|28926365|gb|EAA35342.1|
predicted protein [Neurospora crassa]
Length = 580
Score = 35.4 bits (80), Expect = 1.5
Identities = 31/121 (25%), Positives = 43/121 (34%), Gaps = 10/121 (8%)
Frame = +2
Query: 89 HTHTGGISRNCTLQFVSAVPPQSHTLRARPADRTRGRSEATDSTQARASSCDLHSSSNCG 268
H+ G TLQ ++ PPQS T + + G S +S + SS
Sbjct: 4 HSQLPGRDPRPTLQVITTYPPQSQT--SNYSYPFSGTSPGRESVVSDISSVPSMVEDQGS 61
Query: 269 AVPCGRARQLVHTSANINLWRHRFAHAASHGEVLAT----------HANRCRIPLTPHPH 418
+ Q HTS W + + +H + L T HA RC P TP
Sbjct: 62 DISAEEEYQYDHTSRTRGSWNCFWNNTRAHAKDLQTTDTLARPISHHAGRCTTPSTPERK 121
Query: 419 R 421
R
Sbjct: 122 R 122
>emb|CAD71233.1| hypothetical protein [Neurospora crassa]
Length = 952
Score = 35.0 bits (79), Expect = 1.9
Identities = 24/78 (30%), Positives = 35/78 (44%)
Frame = +3
Query: 720 QAPTTLGYSAHPSSRADTQCSHPSQPDSGSAVQQPHILPTREATSPYLNCGLCTPNTPEH 899
Q +T S+ SS + T + ++ DS SAV P A SP +CG+ TP +
Sbjct: 44 QQTSTSSSSSSSSSSSSTTAATFAKRDSPSAVSTP----ATSAPSPTSSCGVSTPASSTS 99
Query: 900 ACTFSCLTYATGINPTKP 953
+ T TG+ P P
Sbjct: 100 ITANTTTTNLTGVPPIAP 117