Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
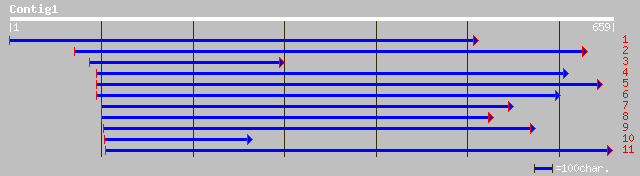
Query= KCC001766A_C01 KCC001766A_c01
(544 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_191640.1| expressed protein [Arabidopsis thaliana] gi|112... 115 5e-25
gb|AAP21391.1| unknown protein [Oryza sativa (japonica cultivar-... 115 5e-25
ref|ZP_00109302.1| COG4446: Uncharacterized protein conserved in... 61 1e-08
ref|NP_485046.1| hypothetical protein [Nostoc sp. PCC 7120] gi|2... 59 5e-08
pir||T43481 probable mucin DKFZp434C196.1 - human (fragment) gi|... 54 1e-06
>ref|NP_191640.1| expressed protein [Arabidopsis thaliana] gi|11290114|pir||T47892
hypothetical protein T4C21.220 - Arabidopsis thaliana
gi|7329691|emb|CAB82685.1| putative protein [Arabidopsis
thaliana]
Length = 214
Score = 115 bits (287), Expect = 5e-25
Identities = 61/145 (42%), Positives = 92/145 (63%), Gaps = 1/145 (0%)
Frame = +3
Query: 111 SARRPTGKGDSEEKL-QPKEIVLRSVNVMVLGALLSIGAAPRPGNLGIIDYGAGVQTLNL 287
SA P D KL ++I+LRS + ++GA+ + + +P LG+ + L L
Sbjct: 28 SASIPETSYDKHPKLIGRRDIILRSSELAMIGAIFQL-SGKKPDYLGVQKN----ERLAL 82
Query: 288 CPPSPNCIATSEEGNDRTHYAPPLTYNPEDGRGKKGPASQEKAMGELVEAVKKLKPDGFT 467
CP + NCI+TSE +DR HYAPP YN G+K P +++ AM EL+ +K +KPD FT
Sbjct: 83 CPATNNCISTSENISDRVHYAPPWNYNG----GRKTPVNRQVAMKELLNVIKSVKPDKFT 138
Query: 468 PKIIKQTDDYLYVEYESPLMGFIDD 542
P+I+++ DDY++VEYESP++G +DD
Sbjct: 139 PRIVEKKDDYVHVEYESPILGLVDD 163
>gb|AAP21391.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|31193901|gb|AAP44736.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 213
Score = 115 bits (287), Expect = 5e-25
Identities = 55/131 (41%), Positives = 85/131 (63%)
Frame = +3
Query: 150 KLQPKEIVLRSVNVMVLGALLSIGAAPRPGNLGIIDYGAGVQTLNLCPPSPNCIATSEEG 329
++ +E VLRS + L A+ + +P LG+ +L LCP + NC++T E+
Sbjct: 37 RIARREFVLRSSELATLAAIFHLSGT-KPRYLGV---QKSPPSLALCPATNNCVSTCEDI 92
Query: 330 NDRTHYAPPLTYNPEDGRGKKGPASQEKAMGELVEAVKKLKPDGFTPKIIKQTDDYLYVE 509
D HYAPP YNP+DGR K P ++ +A+ +L++ V + KPD FTP+++++TDDY+ VE
Sbjct: 93 TDSIHYAPPWNYNPKDGRRAK-PITKHEAINQLIQVVTQTKPDNFTPRLVEKTDDYVRVE 151
Query: 510 YESPLMGFIDD 542
YESP+ GF+DD
Sbjct: 152 YESPIFGFVDD 162
>ref|ZP_00109302.1| COG4446: Uncharacterized protein conserved in bacteria [Nostoc
punctiforme]
Length = 150
Score = 60.8 bits (146), Expect = 1e-08
Identities = 39/121 (32%), Positives = 59/121 (48%)
Frame = +3
Query: 180 SVNVMVLGALLSIGAAPRPGNLGIIDYGAGVQTLNLCPPSPNCIATSEEGNDRTHYAPPL 359
S+ + + G ++ + A RP NLG+ + L CP SPNC+ S + D H PL
Sbjct: 4 SIVLYLPGDVIMVFAGKRPNNLGVSN-----GKLASCPNSPNCV--SSQSADAAHKIAPL 56
Query: 360 TYNPEDGRGKKGPASQEKAMGELVEAVKKLKPDGFTPKIIKQTDDYLYVEYESPLMGFID 539
T+ P A G +K++ KII ++ DYLY E++S L+GF+D
Sbjct: 57 TFT-------SSPQEAMPAAGFAYANLKEIILSLPRTKIITESQDYLYAEFKSALLGFVD 109
Query: 540 D 542
D
Sbjct: 110 D 110
>ref|NP_485046.1| hypothetical protein [Nostoc sp. PCC 7120] gi|25365155|pir||AH1931
hypothetical protein all1003 [imported] - Nostoc sp.
(strain PCC 7120) gi|17130349|dbj|BAB72960.1|
ORF_ID:all1003~hypothetical protein [Nostoc sp. PCC
7120]
Length = 132
Score = 58.5 bits (140), Expect = 5e-08
Identities = 36/104 (34%), Positives = 53/104 (50%)
Frame = +3
Query: 231 RPGNLGIIDYGAGVQTLNLCPPSPNCIATSEEGNDRTHYAPPLTYNPEDGRGKKGPASQE 410
RP NLG+ D L CP SPNC+ S + D H PL + ++ E
Sbjct: 7 RPNNLGVRD-----GRLAPCPNSPNCV--SSQSTDTVHQIAPLNFI----------STPE 49
Query: 411 KAMGELVEAVKKLKPDGFTPKIIKQTDDYLYVEYESPLMGFIDD 542
+A+ +L ++ L KII ++ DYLY E++S L+GF+DD
Sbjct: 50 EAINKLKSVIQSLP----RTKIISESPDYLYAEFQSALLGFVDD 89
>pir||T43481 probable mucin DKFZp434C196.1 - human (fragment)
gi|6599134|emb|CAB63715.1| hypothetical protein [Homo
sapiens]
Length = 580
Score = 53.9 bits (128), Expect = 1e-06
Identities = 58/175 (33%), Positives = 69/175 (39%), Gaps = 22/175 (12%)
Frame = +1
Query: 82 SHCPCGSP-HGLPGGQLARATARRSSSPRRLCSAP*--TLWFWA-RCCPSARPLARATWA 249
S P G+P P G RA A RS S L P +L W R P+ P +
Sbjct: 146 SASPTGTPPRASPTGTPPRAWATRSPSTASLTRTPSRASLTRWPPRASPTRTPPRESPRM 205
Query: 250 SSTTAQACRP*TCALPRPTALPPPRRATTAPTMPLP*PTTPR--MAAARRARPARRRPWA 423
S + PT PP T P P T PR + + RA P R P A
Sbjct: 206 SHRAS------------PTRTPPRASPTRRPPRASPTRTPPRESLRTSHRASPTRMPPRA 253
Query: 424 SWWRRSRSSSPTASPPRSS--------------SRPTTTCT--SSTRARSWGSLT 540
S RR +SPT SPPR+S + PTTT + S TR SW S T
Sbjct: 254 SPTRRPPRASPTGSPPRASPMTPPRASPRTPPRASPTTTPSRASLTRTPSWASPT 308
Score = 44.3 bits (103), Expect = 0.001
Identities = 38/113 (33%), Positives = 48/113 (41%), Gaps = 4/113 (3%)
Frame = +1
Query: 214 PSARPLARATWASSTTAQACRP*TCALPRPTALPPPRRATTAPTMPLP*PTT-PRMAAAR 390
PS L + +S T LPR + + P RA+ T P PT P A+ R
Sbjct: 38 PSRASLKMTPFRASLTKMESTALLRTLPRASLMRTPTRASLMRTPPRASPTRKPPRASPR 97
Query: 391 ---RARPARRRPWASWWRRSRSSSPTASPPRSSSRPTTTCTSSTRARSWGSLT 540
RA P RR P AS +SP +PPR+S T + S T S S T
Sbjct: 98 TPSRASPTRRLPRASPMGSPHRASPMRTPPRASPTGTPSTASPTGTPSSASPT 150
Score = 44.3 bits (103), Expect = 0.001
Identities = 36/112 (32%), Positives = 49/112 (43%)
Frame = +1
Query: 205 RCCPSARPLARATWASSTTAQACRP*TCALPRPTALPPPRRATTAPTMPLP*PTTPRMAA 384
R P A P + AS T P T LPR + + P RA+ T P PT A
Sbjct: 80 RTPPRASPTRKPPRASPRTPSRASP-TRRLPRASPMGSPHRASPMRTPPRASPTGTPSTA 138
Query: 385 ARRARPARRRPWASWWRRSRSSSPTASPPRSSSRPTTTCTSSTRARSWGSLT 540
+ P+ P + R +SPT +PPR+ + + + S TR S SLT
Sbjct: 139 SPTGTPSSASPTGTPPR----ASPTGTPPRAWATRSPSTASLTRTPSRASLT 186
Score = 33.1 bits (74), Expect = 2.5
Identities = 27/79 (34%), Positives = 34/79 (42%), Gaps = 4/79 (5%)
Frame = +1
Query: 316 PPRRATTAPTMPLP*PTTPRMAAARRARP----ARRRPWASWWRRSRSSSPTASPPRSSS 483
P R + T TP MA+ R P R P AS R +S T +PPR+S
Sbjct: 410 PSRASLTRTQSSSSLTRTPSMASLTRTPPRASLTRTPPRASLTRTPPRASLTRTPPRASL 469
Query: 484 RPTTTCTSSTRARSWGSLT 540
T + S R+ S SLT
Sbjct: 470 TRTPSMVSLKRSPSRASLT 488